
miR-203a-3p regulates the cellular processes of esophageal cancer cells via targeting CtBP2
Introduction
Human esophageal cancer (EC) is frequently ranking eighth among all diagnosed cancers all around the world (1-3). It is universally acknowledged that the bad dietary pattern was a main cause of EC, and the incidence of EC was increasing with the patient age (4-6). EC has two main sub-types: esophageal squamous cell cancer (ESCC) and esophageal adeno-cancer (EAC) (7,8). Among them, ESCC is the predominant sub-type of EC, and have a higher risk especially in developing countries (9,10). Despite improvements in chemotherapy, radiotherapy, operation or comprehensive therapeutic strategies, ESCC is often has a poor prognosis and the overall 5-year survival rate of patients usually under 20% on account of metastasis and recurrence (11,12). The molecule mechanism underlying the ESCC tumorigenesis is not yet greatly elucidated. Hence, an in-depth understanding of the initiation and development molecular mechanisms of ESCC is critical important to develop innovative therapeutic strategies for EC (1,13).
MicroRNA (miRNA) is small RNA molecule unable to code, and its length is approximately 19–22 nucleotides (14,15). They can silence downstream target gene expression, in the manner of binding to the 3'-UTR regions of target gene mRNA (16-18). miRNAs are involved in various pathophysiological processes, such as cell growth, cytodifferentiation and apoptosis, in particular, oncogenesis and metastasis (19-21). Besides, miRNAs are dysregulated in many kinds of cancers (such as ESCC) and hence these aberrant miRNAs are good for the diagnosis and therapeutic of cancer patients (22-24). Accumulating evidences demonstrate that, a great deal of tumor suppressive and oncogenic miRNAs participates in ESCC tumorigenesis and tumor development (25). Previous researchers have identified a variety of dysregulated miRNAs in ESCC cells and tissues, including down-regulated in miR-133, miR-187, let-7a, miR-125, miR-377, miR-101 and miR-146 (24,26), as well as up-regulated in miR-210 miR-432-3p, miR-455 and miR-638 and miR-221 (27,28). The previous research demonstrated that miR-203a-3p could suppress cell multiplication and metastases in nasopharyngeal carcinoma through inhibiting LASP1 (29). Further studies showed that PVT1 advanced the progression of ESCC through playing as a molecular sponge of LASP1 and miR-203, offering the best clue of disordered PVT1/miR-203/LASP1 axis in EC (30). Despite numerous miRNAs related with ESCC have been found, the effects of miR-203a-3p on ESCC are currently not fully elaborated.
Previous studies indicated that C-terminal binding protein-2 (CtBP2) played a star role in promoting proliferation and invasion in gastric cancer, breast cancer, prostate cancer, neuroblastoma and ESCC (31-36). Hence, we conceived a strong interest in the upstream regulatory genes of CtBP2. In this study, according to the TargetScan Human 7.1 software which searches for predicted microRNA targets in mammals, we hypothesized that CtBP2 maybe a target gene of miR-203a-3p. So, we designed a list of experiments to verify and further explore the function of miR203a-3p in ESCC in vitro and in vivo.
The research discussed the expression level of miR-203a-3p in ESCC tissues as well as cells, and further clarified the character of miR-203a-3p in ESCC cellular processes via targeting CtBP2 in vitro and in vivo. At the same time, elaborating the underlying molecular mechanisms. Our results demonstrated that miR-203a-3p can be used for an important diagnosis indicator of ESCC and a promising therapeutic target.
Methods
Luciferase reporter assay
To luciferase reporter assay, the CtBP2 recombined vectors were constructed. The diagram of vector construction for luciferase assay was shown in Figure 1A. It was observed that the binding site of miR-203a-3p existed in the CtBP2 3'-UTR. So, the wild type (WT) sequences and mutant type (Mut) sequences of CtBP2 were synthesized and recombined with pGL3 plasmid to construct recombined vector. The pGL3-WT or pGL3-Mut recombined vectors were co-transfected with or without miR-203a-3p negative control (NC) mimic or mimic into HEK293T cells by Lipofectamine 2000 (ThermoFisher Scientific, Waltham, MA, USA). After 48 h transfection, the activity of the luciferase was investigated by the Firefly-Renilla Reporter Assay System kit (BPS Bioscience, San Diego, CA, USA).
Patients and tissue specimens
Ethical approval of this research was authorized by the Committee of Nantong University Affiliated Hospital. To detect the miR-203a-3p expression in ESCC tissues, total 28 patients were recruited in Nantong University Affiliated Hospital during 2010–2017. All diagnoses were confirmed based on histopathological examination. No patients had been received preoperative treatments with radiation therapy or chemotherapeutic agents. After surgical resection, all fresh tissues were immediately thrown into liquid nitrogen, and stored in ultra-low temperature freezer until RNA was isolated. All participants were signed informed consents to use of their tissues for scientific research.
Cell treatment
The ECA109 cell (the human ESCC cell line) and HEK293T were derived from the cell bank (Chinese Academy of Sciences, Shanghai, China). The ECA109 cell and HEK293T were cultured in DMEM (Thermofisher, Waltham, MA) containing with 10 percent fetal cattle serum (FCS) (GE Healthcare Life Sciences, Chicago, IL), at 37 °C in a 5 percent carbon dioxide incubator.
For the in vitro transfection, 2×105/well ECA109 cells were digested and transferred into a 6 well plate and further incubated overnight. under the manufacturer’s advisement, the cells were transfected with 50 nM miR-203a-3p mimic or NC mimic, which were chemical synthesized (RiboBio, Guangzhou, China), by using transfection reagents (RiboBio, Guangzhou, China) separately into each well. The sequences of miRNA mimic were as follows: (sense) 5'-GUGAAAUGUUUAGGACCACUAG-3'; (anti-sense) 5'-CUAGUGGUCCUAAACAUUUCAC-3'. A non-target scrambled miRNA mimic was used as the negative control: (sense) 5'-UUCUCCGAACGUGUCACGU-3'; (anti-sense) 5'-ACGUGACACGUUCGGAGAA-3'. Then cells were collected to verify the miR-203a-3p expression.
RNA isolation and RT-qPCR
Under product instruments, the miRNA was isolated by miRNA Isolation Kit (Qiagen, Hilden, Germany). RNA purity was tested by the 260/280 nm ratio on an OneDrop 1000 spectrophotometer (Wuyi technology. Nanjing, China). RT-qPCR was manipulated triplicate using SYBR Green dye (Chaibio, Santa Clara, CA) on a Lightcycler 96 system (Roche, Basel, Switzerland). 2−∆∆Ct technique was applied to investigate the relative expression of target miRNA and normalized using U6. Bulge-loopTM miRNA qRT-PCR Primer Sets characteristic for miR-203a-3p (RiboBio, Guangzhou, China), and each set had one RT primer and a couple of qRT-PCR primers.
Western blot analysis
Proteins in cells were resolved in lysis buffer which supplemented with 1 mM PMSF, and lysed for 15 min on ice. Briefly, collected the supernatant after centrifugation at 12,000×g for 10 min and diluted in 5×SDS-PAGE loading buffer (Biolegend, San Diego, CA), heated at 95 °C for 5 min and cooled on ice. Protein concentration of each sample was quantified by using of the BCA assay. Then these proteins were transferred onto the PVDF membrane (Bio-Rad, Hercules, CA) after separated. Then the membranes were blocked and reacted with first antibodies against CtBP2, E-cadherin, β-catenin, Vimentin (Biolegend, San Diego, CA) and β-actin (Abcam, Cambridge, MA) in a 4 °C refrigerator overnight. The β-actin was used for the purpose of an internal control. Following, the membranes were reacted with secondary HRP-conjugated antibody. The protein bands were visualized and imaged via chemiluminescence detection system (Tanon, Shanghai, China), and quantified by ImageQuant TL (GE healthcare life sciences, Pittsburgh, PA).
MTT
After the ECA109 cells were treated with 50 nM NC mimic or miRNA mimic transfection for 48 h, Cells were digested and transferred into 96-well plates at a 104 cells/well. Added 10 µL MTT (5 mg/mL) per well after inoculated for 24 h at 37 °C, and followed by further culture for 4 h. And 100 µL each well of DMSO was put into dissolve the MTT crystals. TECAN infinite F200 (TECAN, Switzerland) was chosen to read the absorbance values at 570 nm.
Transwell and wound healing assay
Transwell chambers (CORNING, Corning, NY) were chosen to explore the impact of miR-203a-3p on cell migration and invasion. After transfection, 1×105 cells were resuspended in 200 µL DMEM and then transplanted into upper chambers in 24-well plates. Prior to seed cells, the upper surface of chambers was pre-coated using 50 µL Matrigel before the invasion assay. After transfection 24 h, cells that did not pass through the membrane were cautiously scrubbed by cotton swabs. And the cells transferred to the basement of the membranes were fixed with 4% PFA, and dyed with 0.1% crystal violet (Abcam, Cambridge, UK) for 40 min. The number of migrating and invading cells were photographed and counted 5 randomly selected fields by using a phase contrast microscope (×200) (Zeiss, Oberkochen, Germany).
About the wound healing experiment, seeded 1×105 transected cells per well into 24-well plates, then subcultured to form a monolayer. When the cell density reached over 90%, the monolayer was scratched by using a 100 µL pipette. Then washed the cells twice with DMEM and appended fresh serum-free medium to further incubation. Photos were taken randomly with microscopy (Zeiss, Oberkochen, Germany) at 0 h, 24 h and 48 h. Wound healing rate (%) = (x h Scratch area − 0 h Scratch area)/0 h Scratch area ×100.
TUNEL assay
The ECA109 cells were digested and transplanted into 24-well plates and cultured overnight after transfection. Under the user manual, TUNEL positive cells were detected by using of a TUNEL Kit (Biovision, Milpitas, CA). Firstly, cells were fixed with 4 percent PFA for 20 min at normal temperature and then washed with PBS. Secondly, the fixed cells were incubated with blocking solution for 10 min and then strengthened the permeability with permeabilization solution. Thirdly, added an addition of 50 µL TUNEL reaction solution per well and reacted at 37 °C for 1 h in a humidifying container. Finally, the nucleus was dyed with 1 µg/mL Hoechst 33258. The images were acquired under a fluorescence microscopy in randomly three fields each slice. Image J was applied to count and analyze the TUNEL positive cells. Apoptotic rate = TUNEL positive cells/total cells ×100%.
Flow cytometry (FCM) assay
For cell proliferation assay, cell cycle was assayed with FCM (BD Bioscience, San Jose, CA). Firstly, transfected ECA109 cells were digested with trypsin, and rinsed with PBS. Secondly, the cells at 37 °C were reacted with 500 µL PI (Biotium, Fremont, CA) mixture solution protected from light for 30 min. Finally, the cell percentage in cell cycle was analyzed by FCM.
Annexin V staining and FCM assay was applied to investigate apoptosis. According to the Annexin V-FITC Kit (Biotium, Fremont, CA) protocol, the cells after transfection for 48 h in each group were digested with 0.25% typsin and rinsed with PBS. Cells were reacted with 5 µL Annexin V-FITC, 10 µL PI diluted in 195 µL binding buffer mixture solution for 10 min protected from light. FCM (BD Pharmingen, San Diego, CA, USA) was chosen to measure cell apoptosis of ECA109 cells. 10,000 cells with fluorescence signals of each group were gated, and analyzed the data by Macquit software.
Rescue experiment
For rescue experiment, the recombined lentivirus vector (LV-CtBP2+) has been constructed to overexpression of CtBP2 (33). In rescue experiment, the recombined lentivirus vector (LV-CtBP2+) and miR-203a-3p was co-transfected into ECA109 cells.
Xenograft tumor model and in vivo metastasis
3.5×106 ECA 109 cells/100 µL each mouse were subcutaneously injected into 10 male, 6 weeks old immunodeficient mice (BALB/c nude) (Laboratory Animal Center of Yangzhou University, Yangzhou, China). After 7 days, they were randomly divided into two groups (n=5): agomir NC group and agomir group. The mice were injected intratumorally with miR-203a-3p agomir or agomir NC (RiboBio Co., Ltd., Guangzhou, China) at a dose of 1 nmol per mouse every 3 days. To track the growth of tumor in vivo, we measured the tumor volume with calipers every 3 days for 24 days. Tumor volume was counted as follow: (length × width2)/2. Subsequently, tumors volume was measured to compare the tumor growth rate in each group. Animal experiments were approved by the Nantong University Animal Care and Use Committee.
Statistical analysis
All data were demonstrated as the mean ± standard error of mean and statistical analysis was carried on by SPSS software. Comparison of two groups was analyzed by ANOVA and t-test.
Results
CtBP2 was a direct target gene of miR-203a-3p
MiRNAs generally regulate their target gene by directly binding to their 3'-UTR.
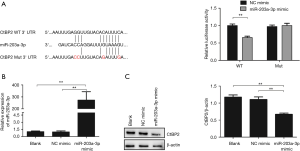
Based on the analytical result of the TargetScan Human 7.1 software (http://www.targetscan.org), we found that 3'-UTR of CtBP2 mRNA harbors an AU-rich element, which is the potential binding site of miR-203a-3p (Figure 1A). In order to verify the CtBP2 was a direct target of miR-203a-3p, luciferase assay system was adopted. So, we constructed two luciferase recombined vectors (pGL3-WT and pGL3-Mut), which containing the wild WT and Mut sequences in 3'-UTR of CtBP2. After the HEK293T cell was co-transfected with miR-203a-3p mimic and pGL3-WT or pGL3-Mut, the relative luciferase activity was measured. The results displayed that the relative luciferase activity in the group treated with miR-203a-3p mimic and pGL3-WT co-transfection was remarkably decreased (P<0.01), while barely decreased in miR-203a-3p mimic and pGL3-CtBP2-3'-UTR-Mut group (Figure 1A).
Then, RT-qPCR and Western blot was chosen to further test the impact of miR-203a-3p on CtBP2 expression. The results of RT-qPCR revealed that the miR-203a-3p expression was dramatically increased in miR-203a-3p mimic group, compared to blank and NC mimic groups (P<0.01) (Figure 1B). As we expected, the CtBP2 expression was markedly reduced in miR-203a-3p mimic group (P<0.01) (Figure 1C). The above results indicated that miR-203a-3p could inhibit specifically CtBP2 expression.
miR-203a-3p inhibited the proliferation, migration and invasion, while promoted apoptosis in ECA109 cells
Our previous research revealed that the CtBP2 expression was markedly raised in ESCC tissues (32,37). In order to further determine the character of miR-203a-3p in ESCC, we surveyed the miR-203a-3p expression in ESCC tissues and peri-neoplastic tissues by RT-qPCR. The miR-203a-3p expression in ESCC tissues and cells was dramatically decreased, compared to peri-neoplastic tissues (P<0.01) (Figure 2A).
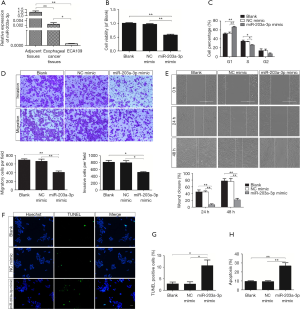
CtBP2 played a role similar to oncogene in tumorigenesis and development. Aim to test the character of miR-203a-3p in the cellular process of ECA109 cell, MTT, transwell, wound healing, TUNEL and FCM assay was adopted. Firstly, MTT assay was applied to test the impact of miR-203a-3p on the cell viability. The ECA109 cell viability in miR-203a-3p mimic group was dropped significantly (P<0.01) (Figure 2B). So, miR-203a-3p overexpression could reduce cell vitality.
Secondly, as showed in the result of FCM assay, cells in miR-203a-3p mimic group get arrested obviously in S-phase (Figure 2C). The result reflected that overexpression of miR-203a-3p could retard the cell proliferation in ECA109 cells.
Thirdly, transwell assay was applied to test the impact of miR-203a-3p on the migration and invasion of ECA109 cells. The cell numbers of miR-203a-3p mimic transfection group were significantly reduced (P<0.05) (Figure 2D). So, miR-203a-3p overexpression remarkably restrained both cell migration and invasion in ECA109 cells. Moreover, wound healing assay clarified that the miR-203a-3p overexpression reduced cell motility (P<0.01) (Figure 2E). The results of wound healing assay were similar to the transwell assay. Overall, up-regulation of miR-203a-3p dramatically inhibited migration and invasion of the ECA109 cell.
TUNEL assay and FCM assay were adopted to further ascertain the impact of miR-203a-3p on apoptosis. The percentage of TUNEL positive cells in miR-203a-3p mimic group was 10.7±3.4, which was markedly raised (P<0.05) (Figure 2F,G). As shown in Figure 2H, the results of FCM assay were accordance with TUNEL staining, miR-203a-3p mimic group had a higher apoptosis rate than that in blank group or NC mimic group (P<0.01). Above outcomes showed that miR-203a-3p overexpression could accelerate the apoptosis in ECA109 cells.
miR-203a-3p restrained epithelial-mesenchymal transition (EMT) in ECA109 cells
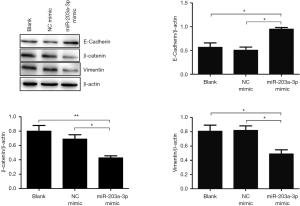
EMT is an essential process for the initiation of metastasis in cancer. To explore whether miR-203a-3p restrained the metastasis of ECA109 cells by depressing EMT, we measured the expression of EMT biomarkers using Western blot. So, the three proteins (E-cadherin, β-catenin and vimentin) involved in EMT were tested. The expressions of E-cadherin and β-catenin in miR-203a-3p transfection group were significantly increased (P<0.05), while the vimentin expression was significantly dropped (P<0.05) (Figure 3). The results indicated that miR-203a-3p overexpression restrained EMT via up-regulating E-cadherin and β-catenin, and down-regulating vimentin in ECA109 cells.
miR-203a-3p regulated the cellular processes of ECA109 cells via targeting CtBP2
The above results pointed out that CtBP2 is the direct target gene of miR-203a-3p. So, we employed rescue experiment to further verify the function of miR-203a-3p in the cellular processes via directly targeting CtBP2. For rescue experiment, the recombined lentivirus vector (LV-CtBP2+) has been constructed to overexpression of CtBP2 (33). Because of the recombined vector with Flag, we tested the expression of Flag to measure the CtBP2 expression by Western blot. The expression of Flag in the LV-CtBP2+ transfection group was dramatically increased (Figure 4A). So, the CtBP2 stable overexpression cell line was used in the following rescue experiment. As anticipated, the expression of Flag in (LV-CtBP2+ + miR-203a-3p mimic) group was markedly reduced (P<0.05), compared to LV-CtBP2+ and (LV-CtBP2+ + NC mimic) group (Figure 4A).
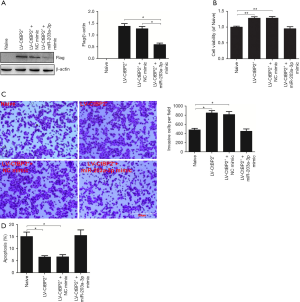
Next, we tested impact of miR-203a-3p on the cell viability via targeting CtBP2 by MTT. It turned out that cell vitality was improved in LV-CtBP2+ group, compared with blank group (P<0.01). Surprisingly, after cell was co-treated with miR-203a-3p mimic and LV-CtBP2+ for 48 h, the viability of ECA109 cells was decreased. And the cell viability in naive and (LV-CtBP2+ + miR-203a-3p mimic) groups was so close (P>0.05) (Figure 4B). CtBP2 overexpression reversed the impact of miR-203a-3p on the cell vitality in ECA109 cells.
Given the evidence of miR-203a-3p significantly inhibited the metastasis of ECA109 cell, we then further assessed miR-203a-3p regulated the invasion of ECA109 cell via targeting CtBP2 by transwell assay. As we expected, the cell numbers of group treated with LV-CtBP2+ transfection were markedly increased (P<0.05), and the cell numbers in the naive group and (LV-CtBP2+ + miR-203a-3p mimic) group was almost equal (P>0.05) (Figure 4C). These outcomes suggested that CtBP2 overexpression reversed the impact of miR-203a-3p on the cell invasion in ECA109 cell.
FCM assay was used to assess whether miR-203a-3p regulated ECA109 cell apoptosis via targeting CtBP2. Unsurprisingly, the apoptotic rate in LV-CtBP2+ transfection group was evidently reduced (P<0.05), and the apoptotic rate in the naive group and (LV-CtBP2+ + miR-203a-3p mimic) group was almost at the same level (P>0.05) (Figure 4D). Overall, overexpression of miR-203a-3p reversed decreased apoptosis rate induced by CtBP2 overexpression.
In summary, these outcomes powerfully proved that miR-203a-3p regulated the cellular processes of ECA109 cells via targeting CtBP2.
miR-203a-3p suppressed tumor growth in vivo
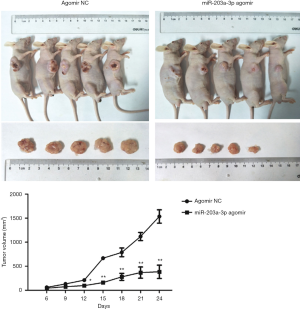
ECA109 cells were subcutaneously injected into the immunodeficient mice. We measured the tumor volume with calipers every 3 days for 24 days. The results revealed that tumor volumes of the mice in miR-203a-3p agomir group were remarked atrophied (Figure 5).
Discussion
ESCC is the major histological type of EC in China with a poor prognosis (10,38). With improvements in strategies, the mortality of ESCC have decreased remarkably, whereas the survival rate was still not optimistic which result from high recurrence and distant metastasis rates after surgery (39-41). However, the molecular mechanism underlying the tumorigenesis and metastasis of ESCC remains greatly unknown. Some reports presented that numerous carcinogenic and anticancer miRNAs participated in ESCC progression and tumor development via modulating cellular processes (14,42,43). miR-203a-3p was participated in many types of tumors such as liver cancer, nasopharyngeal carcinoma and hepatocellular carcinoma (29,44,45). Among these cancers the miR-203a-3p expression was down-regulated, revealing it maybe acts as a tumor suppressor.
CtBP expression was remarkedly raised in several kinds of neoplasm tissues and cell lines compared to peri-neoplastic tissues, such as ESCC. In the previous studies showed that CtBP2 involved in tumorigenesis, progression and Cisplatin chemoresistance in ESCC (32,33). These results demonstrated that CtBP2 played an oncogene role in ESCC. Aim to investigate the upstream genes of CtBP2, we assumed that miRNA could regulate CtBP2. Based on predicted result of the TargetScan Human 7.1 software, CtBP2 was possible a target gene of miR-203a-3p by directly binding to its 3'-UTR. Firstly, the prediction was identified by luciferase reporter assay. As we expected, CtBP2 was a direct target gene of miR-203a-3p and its expression was regulated directly by miR-203a-3p.
Secondly, a series of methods were designed to check the impact of miR-203a-3p on the cellular processes in ECA109 cells. These outcomes revealed that miR-203a-3p could restrain cell proliferation, migration and invasion, and accelerate apoptosis. EMT is an essential process for the initiation of metastasis in cancer (46). Moreover, miR-203a-3p could restrain EMT in ECA109 cells. These results indicated that miR-203a-3p may be played an antioncogene role in ESCC. Next, rescue experiment was used to further confirm that miR-203a-3p regulated the cellular processes in ECA109 cells via targeting CtBP2. Unsurprisingly, overexpression of CtBP2 could reverse the impact of miR-203a-3p on the cellular processes in ECA109 cells.
Thirdly, most interesting, we constructed a xenograft tumor model in immunodeficient mice to test the influence of miR-203a-3p on the neoplasm growth in vivo. Amazing, miR-203a-3p significantly suppress the growth of xenograft tumor in nude mice. These outcomes revealed that miR-203a-3p maybe have a wide range of application in the therapy of ESCC in the future.
Conclusions
In conclusion, our results ascertained that miR-203a-3p was down-regulated in ESCC tissues and it could inhibit cell proliferation, migration and invasion, and promote apoptosis in vitro by negatively regulating CtBP2. Moreover, miR-203a-3p could apparently suppress tumor growth in vivo. Therefore, the outcomes revealed that miR-203a-3p can be used as a diagnosis indicator and a potential therapeutic target for ESCC.
Acknowledgments
Funding: This study was supported by grants from
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.44). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Committee of Nantong University Affiliated Hospital. All participants were signed informed consents to use of their tissues for scientific research. Animal experiments were approved by the Nantong University Animal Care and Use Committee in compliance with the institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Steins A, Ebbing EA, Creemers A, et al. Chemoradiation induces epithelial-to-mesenchymal transition in esophageal adenocarcinoma. Int J Cancer 2019;145:2792-803. [Crossref] [PubMed]
- Shi H, Shi J, Zhang Y, et al. Long non-coding RNA DANCR promotes cell proliferation, migration, invasion and resistance to apoptosis in esophageal cancer. J Thorac Dis 2018;10:2573-82. [Crossref] [PubMed]
- Athanasiou A, Spartalis M, Spartalis E. Hybrid Minimally Invasive Esophagectomy for Esophageal Cancer. N Engl J Med 2019;380:e28. [Crossref] [PubMed]
- Deng J, Chen H, Zhou D, et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat Commun 2017;8:1533. [Crossref] [PubMed]
- Li C, Wang X, Wang X, et al. A multicenter phase III study comparing Simultaneous Integrated Boost (SIB) radiotherapy concurrent and consolidated with S-1 versus SIB alone in elderly patients with esophageal and esophagogastric cancer - the 3JECROG P-01 study protocol. BMC Cancer 2019;19:397. [Crossref] [PubMed]
- Doi T, Piha-Paul SA, Jalal SI, et al. Safety and Antitumor Activity of the Anti-Programmed Death-1 Antibody Pembrolizumab in Patients With Advanced Esophageal Carcinoma. J Clin Oncol 2018;36:61-7. [Crossref] [PubMed]
- Deswysen Y, Van Ginderachter L, Dano H, et al. Mimicking oesophageal cancer: oesophageal actinomycosis. Lancet Infect Dis 2019;19:558. [Crossref] [PubMed]
- Blum Murphy M, Xiao L, Patel VR, et al. Pathological complete response in patients with esophageal cancer after the trimodality approach: The association with baseline variables and survival-The University of Texas MD Anderson Cancer Center experience. Cancer 2017;123:4106-13. [Crossref] [PubMed]
- Nie C, Qin X, Li X, et al. CACNA2D3 Enhances the Chemosensitivity of Esophageal Squamous Cell Carcinoma to Cisplatin via Inducing Ca(2+)-Mediated Apoptosis and Suppressing PI3K/Akt Pathways. Front Oncol 2019;9:185. [Crossref] [PubMed]
- Yan T, Cui H, Zhou Y, et al. Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma. Nat Commun 2019;10:1670. [Crossref] [PubMed]
- Newton AD, Predina JD, Xia L, et al. Surgical Management of Early-Stage Esophageal Adenocarcinoma Based on Lymph Node Metastasis Risk. Ann Surg Oncol 2018;25:318-25. [Crossref] [PubMed]
- Su Z, Zou GR, Mao YP, et al. Prognostic impact of family history of cancer in Southern Chinese patients with esophageal squamous cell cancer. J Cancer 2019;10:1349-57. [Crossref] [PubMed]
- Neuber B, Schmitt M. Engineered natural killer cells expressing chimeric antigen receptors (CAR)—a promising approach in tumor immunotherapy. Biotarget 2019;3:1. [Crossref]
- Scheunemann D, Pradhan AK, Das SK, et al. Wnt7a and miR-370-3p: new contributors to bladder cancer invasion. Biotarget 2018;2:14. [Crossref] [PubMed]
- Starlinger P, Hackl H, Pereyra D, et al. Predicting Postoperative Liver Dysfunction Based on Blood Derived MicroRNA Signatures. Hepatology 2019;69:2636-51. [PubMed]
- Qiao L, Hu S, Liu S, et al. MicroRNA-21-5p dysregulation in exosomes derived from heart failure patients impairs regenerative potential. J Clin Invest 2019;130. [PubMed]
- Karimi L, Zeinali T, Hosseinahli N, et al. miRNA-143 replacement therapy harnesses the proliferation and migration of colorectal cancer cells in vitro. J Cell Physiol 2019;234:21359-68. [Crossref] [PubMed]
- Pandolfini L, Barbieri I, Bannister AJ, et al. METTL1 Promotes let-7 MicroRNA Processing via m7G Methylation. Mol Cell 2019;74:1278-90.e9. [Crossref] [PubMed]
- Crosby P, Hamnett R, Putker M, et al. Insulin/IGF-1 Drives PERIOD Synthesis to Entrain Circadian Rhythms with Feeding Time. Cell 2019;177:896-909.e20. [Crossref] [PubMed]
- Zhang T, Xue X, Peng H. Therapeutic Delivery of miR-29b Enhances Radiosensitivity in Cervical Cancer. Mol Ther 2019;27:1183-94. [Crossref] [PubMed]
- Wang X, Chen Q, Yi S, et al. The microRNAs let-7 and miR-9 down-regulate the axon-guidance genes Ntn1 and Dcc during peripheral nerve regeneration. J Biol Chem 2019;294:3489-500. [Crossref] [PubMed]
- Gao Y, Yi J, Zhang K, et al. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J Exp Clin Cancer Res 2017;36:161. [Crossref] [PubMed]
- Lindner K, Eichelmann AK, Matuszcak C, et al. Complex Epigenetic Regulation of Chemotherapy Resistance and Biohlogy in Esophageal Squamous Cell Carcinoma via MicroRNAs. Int J Mol Sci 2018; [Crossref] [PubMed]
- Li B, Xu WW, Han L, et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene 2017;36:3986-4000. [Crossref] [PubMed]
- Shimonosono M, Idichi T, Seki N, et al. Molecular pathogenesis of esophageal squamous cell carcinoma: Identification of the antitumor effects of miR1453p on gene regulation. Int J Oncol 2019;54:673-88. [PubMed]
- Jamali L, Tofigh R, Tutunchi S, et al. Circulating microRNAs as diagnostic and therapeutic biomarkers in gastric and esophageal cancers. J Cell Physiol 2018;233:8538-50. [Crossref] [PubMed]
- Ma J, Zhan Y, Xu Z, et al. ZEB1 induced miR-99b/let-7e/miR-125a cluster promotes invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett 2017;398:37-45. [Crossref] [PubMed]
- Liu SG, Qin XG, Zhao BS, et al. Differential expression of miRNAs in esophageal cancer tissue. Oncol Lett 2013;5:1639-42. [Crossref] [PubMed]
- Jiang N, Jiang X, Chen Z, et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res 2017;36:138. [Crossref] [PubMed]
- Li PD, Hu JL, Ma C, et al. Upregulation of the long non-coding RNA PVT1 promotes esophageal squamous cell carcinoma progression by acting as a molecular sponge of miR-203 and LASP1. Oncotarget 2017;8:34164-76. [PubMed]
- Zhao Z, Hao D, Wang L, et al. CtBP promotes metastasis of breast cancer through repressing cholesterol and activating TGF-beta signaling. Oncogene 2019;38:2076-91. [Crossref] [PubMed]
- Guan C, Shi H, Wang H, et al. CtBP2 contributes to malignant development of human esophageal squamous cell carcinoma by regulation of p16INK4A. J Cell Biochem 2013;114:1343-54. [Crossref] [PubMed]
- Shi H, Mao Y, Ju Q, et al. C-terminal binding protein2 mediates cisplatin chemoresistance in esophageal cancer cells via the inhibition of apoptosis. Int J Oncol 2018;53:167-76. [PubMed]
- Dai F, Xuan Y, Jin JJ, et al. CtBP2 overexpression promotes tumor cell proliferation and invasion in gastric cancer and is associated with poor prognosis. Oncotarget 2017;8:28736-49. [PubMed]
- Nan J, Guan S, Jin X, et al. Down-regulation of C-terminal binding protein 2 (CtBP2) inhibits proliferation, migration, and invasion of human SHSY5Y cells in vitro. Neurosci Lett 2017;647:104-9. [Crossref] [PubMed]
- Xuan Q, Zhong X, Li W, et al. CtBP2 is associated with angiogenesis and regulates the apoptosis of prostate cancer cells. Oncol Rep 2017;38:1259-67. [Crossref] [PubMed]
- Zhang J, Zhu J, Yang L, et al. Interaction with CCNH/CDK7 facilitates CtBP2 promoting esophageal squamous cell carcinoma (ESCC) metastasis via upregulating epithelial-mesenchymal transition (EMT) progression. Tumour Biol 2015;36:6701-14. [Crossref] [PubMed]
- Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol 2017;27:215-21. [Crossref] [PubMed]
- Liu CC, Chou KT, Hsu JW, et al. High metabolic rate and stem cell characteristics of esophageal cancer stem-like cells depend on the Hsp27-AKT-HK2 pathway. Int J Cancer 2019;145:2144-56. [Crossref] [PubMed]
- Wang Z, Kang L, Zhang H, et al. AKT drives SOX2 overexpression and cancer cell stemness in esophageal cancer by protecting SOX2 from UBR5-mediated degradation. Oncogene 2019;38:5250-64. [Crossref] [PubMed]
- Liu X, Song M, Wang P, et al. Targeted therapy of the AKT kinase inhibits esophageal squamous cell carcinoma growth in vitro and in vivo. Int J Cancer 2019;145:1007-19. [Crossref] [PubMed]
- Bahmanpour Z, Sheervalilou R, Choupani J, et al. A new insight on serum microRNA expression as novel biomarkers in breast cancer patients. J Cell Physiol 2019;234:19199-211. [Crossref] [PubMed]
- Macharia LW, Wanjiru CM, Mureithi MW, et al. MicroRNAs, Hypoxia and the Stem-Like State as Contributors to Cancer Aggressiveness. Front Genet 2019;10:125. [Crossref] [PubMed]
- Huo W, Du M, Pan X, et al. miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma cell growth and metastasis. FEBS Open Bio 2017;7:1085-91. [Crossref] [PubMed]
- Wang L, Tong D, Guo Q, et al. HOXD3 targeted by miR-203a suppresses cell metastasis and angiogenesis through VEGFR in human hepatocellular carcinoma cells. Sci Rep 2018;8:2431. [Crossref] [PubMed]
- Aiello NM, Kang Y. Context-dependent EMT programs in cancer metastasis. J Exp Med 2019;216:1016-26. [Crossref] [PubMed]


