
Reduced RANBP9 expression is associated with poor prognosis in colorectal cancer patients
Introduction
Colorectal cancer (CRC) is the third most common malignancy worldwide. It has a poor prognosis, with 5 and 10-year relative survival rates of 65% and 58%, respectively (1). TNM staging is the gold standard for CRC staging and is used to evaluate tumor progression and as a guide to select suitable therapy (2). However, there are significant differences in prognosis even for patients with the same tumor stage (3,4). This suggests that traditional TNM staging does not consider all the biological characteristics of the tumor. Hence, new reliable prognostic factors are required.
Several genetic and molecular changes accumulate during CRC development. For example, most CRCs have chromosomal instability, leading to loss of heterozygosity at various tumor suppressor gene loci or mutations in oncogene loci. This is followed by microsatellite instability and DNA promoter hypermethylation (5,6). Mutations in BRAF, NRAS and KRAS affect patient prognosis receiving chemotherapy or targeted therapy (7,8). They have been used to evaluate prognosis or responsiveness to treatment (5,9). However, molecular CRC typing does not have a unified standard. Hence, identifying additional molecular markers is critical for proper patient prognosis.
RANBP9 is a RAN-binding protein, and participates in nucleation of the centriole microtubules, and affects cell division and differentiation (10). It has been studied in nerve tissues, immune system, and cancer. In breast cancer, its expression was found to correlate with specific clinical features and inflammatory cell infiltrates within the tumor (11). It is highly expressed in osteosarcoma and suppresses metastasis by accelerating mitochondrial-associated anoikis (12,13). In gastric cancer, reduced RANBP9 expression is associated with distant metastasis and chemoresistance (14). In lung cancer, reduced RANBP9 mRNA expression levels are associated with poor survival and increased proliferation and invasion in vitro (15). RANBP9 protein levels have been reported to be upregulated in lung tumors, and is negatively correlated with outcome and response to DNA damaging agents (16,17). However, the mechanism of RANBP9 in CRC is yet to be deciphered.
In our previous study, we found that RANBP9 was overexpressed in CRC tissues in a small patient cohort. It suppressed CRC cell proliferation both in vitro and in vivo (18). The aim of this study was to determine the association between RANBP9 expression levels and clinicopathological parameters in an expanded patient cohort, as well as CRC patient prognosis. Reduced RANBP9 expression was found to be correlated with vascular invasion, tumor location, normal serum carcinoembryonic antigen (CEA), and lower survival rates following radical resection. Hence, RANBP9 could be an independent prognostic factor.
Methods
Patients and tissue samples
Consecutive CRC patients (aged from 18 to 75 years), who underwent radical resection (both primary lesion and metastases, if present) at Jinshan Hospital of Fudan University (Shanghai, China) between January 1st, 2012, and February 28th, 2013, were selected for this study. Patients who underwent neoadjuvant chemotherapy were excluded from this study. Selected CRC patients were TNM classified based on the seventh edition of the American Joint Committee on Cancer (AJCC) TNM staging system (2). All patients were followed-up in the same hospital every 3 months for the first two years and every 6 months afterwards. We reviewed medical records in order to assess clinicopathological characteristics such as age, gender, tumor location, tumor differentiation, vascular invasion, perineural invasion, TNM stages, pre-surgical serum CEA and post-surgical chemotherapy. Disease-free survival (DFS) was defined as the time of surgery to the date of disease recurrence or death from any cause. Overall survival (OS) was defined as the interval between the date of surgery and mortality due to any cause. Ethical approval was obtained from the Ethics Committee of Jinshan Hospital, Fudan University (Shanghai, China, No. 2011-02-01). Written informed consents for the use of tissue samples was obtained from all patients.
Immunohistochemistry (IHC)
Tissue samples were used for IHC analysis and were performed as previously described (19). Briefly, slides were blocked with goat serum (Haoranbio, Shanghai, China), and incubated with polyclonal anti-RANBP9 antibody (1:100; cat. no. 17755-1-AP; Proteintech, Wuhan, Hubei, China). Afterwards the slides were incubated with a secondary antibody (1:2000; cat. no. KGAA35; KeyGen Biotech, Nanjing, Jiangsu, China). The RANBP9 antibody was validated for specificity in human samples in our previous study (18). All slides were observed using the Olympus BX43 (Tokyo, Japan) microscope, and evaluated by two investigators who were blinded to the clinicopathological characteristics of the study participants. In case of discrepancy, consensus was always achieved after discussion. IHC scoring for RANBP9 expression was performed using a semi-quantitation method as previously described, and was based on the multiplication of intensity and percentage of stained cells (20). RANBP9 expression intensity was scored from 0 to 3 (0, negative; 1, weak; 2, moderate; 3, strong). The percentage of positively stained cells was scored from 0 to 4 (1, 0–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%). Three sections of each tumor were assessed for RANBP9 staining and the average score was calculated. Based on the IHC scores, RANBP9 staining patterns were classified as low-level (IHC score, 0 to 4) or high-level (IHC score, 6 to 12).
Statistical analysis
Data was analyzed using SPSS 23.0 software (IBM, Armonk, NY, USA). Comparison between RANBP9 expression levels and clinicopathological CRC parameters was performed using a χ2 test. DFS and OS curves were estimated using the Kaplan-Meier method and compared using log-rank test. Cox regression analysis was used to summarize the clinicopathological characteristics with respect to DFS and OS. All P values were two-sided and considered statistically significant if less than 0.05.
Results
Clinical characteristics of the CRC patients
The 228 CRC patients were between 28 to 75 years old (median, 59 years), of which, 155 (68.0%) were male and 73 (32.0%) were female (Table 1). The primary tumor site was right-sided for 86 (37.7%) of the patients and left-sided for 142 (62.3%) of the patients. Based on the depth of tumor invasion, 52 (22.8%) patients were staged at T1 and T2, while 176 (77.2%) patients were staged at T3 and T4. Lymph node metastases were present in 98 (43.0%) patients. Distant metastasis was present in seven (3.1%) patients whose tumors were radically resected, and were included in the TNM III, VI subclass. Nine patients were lost during follow-up and were excluded. The cutoff date for survival data was December 31st, 2017. The median follow-up time for the 228 CRC patients was 61 months.
Table 1
| Characteristics | n (%) |
|---|---|
| Age, years | |
| <65 | 122 (53.5) |
| ≥65 | 106 (46.5) |
| Gender | |
| Male | 155 (68.0) |
| Female | 73 (32.0) |
| Tumor location | |
| Right colon | 86 (37.7) |
| Left colon | 142 (62.3) |
| Differentiation | |
| Well | 116 (50.9) |
| Poorly | 112 (49.1) |
| Vascular invasion | |
| Present | 19 (8.3) |
| Absent | 209 (91.7) |
| Perineural invasion | |
| Present | 12 (5.3) |
| Absent | 216 (94.7) |
| T stage | |
| T1, 2 | 52 (22.8) |
| T3, 4 | 176 (77.2) |
| N stage | |
| N0 | 130 (57.0) |
| N1, 2 | 98 (43.0) |
| TNM stages | |
| I, II | 127 (55.7) |
| III, IV | 101 (44.3) |
| Serum CEA | |
| Normal | 95 (41.7) |
| Elevated | 133 (58.3) |
| Post-surgical chemotherapy | |
| No | 54 (23.7) |
| Yes | 174 (76.3) |
TNM, tumor node metastasis, CEA, carcinoembryonic antigen.
Association between RANBP9 expression levels and CRC clinicopathological parameters
IHC was used to measure RANBP9 expression levels in paraffin-embedded tissue samples from CRC patients (Figure 1). Patient samples were divided into low RANBP9 expression (n=119) and high RANBP9 expression (n=109) groups based on RANBP9 staining. Clinicopathological parameters are summarized in Table 2. Low RANBP9 expression levels were correlated with tumor location (P=0.014), vascular invasion (P=0.057, marginally significant) and normal serum CEA levels (P=0.001). No statistically significant association was observed for other parameters, including age, gender, differentiation, perineural invasion, TNM stage and postoperative chemotherapy.
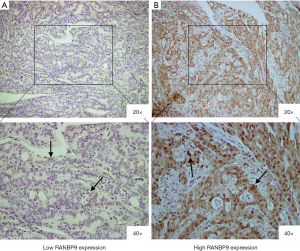
Table 2
| Characteristic | RANBP9 expression intensity | P value* | |
|---|---|---|---|
| Low (%) | High (%) | ||
| Age, years | 0.288 | ||
| <65 | 68 (29.8) | 54 (23.7) | |
| ≥65 | 51 (22.4) | 55 (24.1) | |
| Gender | 0.572 | ||
| Male | 83 (36.4) | 72 (31.6) | |
| Female | 36 (15.8) | 37 (16.2) | |
| Tumor location | 0.014 | ||
| Right colon | 54 (23.7) | 32 (14.0) | |
| Left colon | 65 (28.5) | 77 (33.8) | |
| Differentiation | 0.236 | ||
| Well | 56 (24.6) | 60 (26.3) | |
| Poorly | 63 (27.6) | 49 (21.5) | |
| Vascular invasion | 0.057 | ||
| Present | 14 (6.1) | 5 (2.2) | |
| Absent | 105 (46.1) | 104 (45.6) | |
| Perineural invasion | 1.000 | ||
| Present | 6 (2.6) | 6 (2.6) | |
| Absent | 113 (49.6) | 103 (45.2) | |
| T stage | 0.636 | ||
| T1, 2 | 29 (12.7) | 23 (10.1) | |
| T3, 4 | 90 (39.5) | 86 (37.7) | |
| N stage | 0.349 | ||
| N0 | 64 (28.1) | 66 (28.9) | |
| N1, 2 | 55 (24.1) | 43 (18.9) | |
| TNM stages | 0.424 | ||
| I, II | 63 (27.6) | 64 (28.1) | |
| III, IV | 56 (24.6) | 45 (19.7) | |
| Serum CEA | 0.001 | ||
| Normal | 62 (27.2) | 33 (14.5) | |
| Elevated | 57 (25.0) | 76 (33.3) | |
| Post-surgical chemotherapy | 0.876 | ||
| No | 29 (12.7) | 25 (11.0) | |
| Yes | 90 (39.5) | 84 (36.8) | |
| Total | 119 (52.2) | 109 (47.8) | |
*Differences between the two groups were evaluated using χ2 test. RANBP9, RAN-binding protein 9. TNM, tumor node metastasis, CEA, carcinoembryonic antigen.
Survival analysis based on RANBP9 expression levels
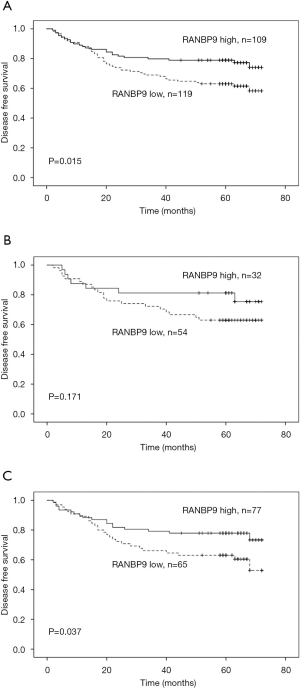
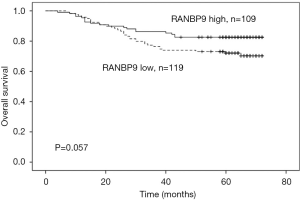
Of the 228 selected patients, 71 (31.1%) had an event during the follow-up period; there were 46 patients with low RANBP9 expression levels and 25 patients with high RANBP9 expression levels. Kaplan-Meier curves and log-rank tests were used to determine the association between RANBP9 expression levels and DFS rates. Patients with low RANBP9 expression levels had a 5-year DFS rate of 63.0% compared to 78.9% of patients with high RANBP9 expression levels (P=0.015; Figure 2A). Subgroup analysis of the prognostic value of RANBP9 expression levels was performed for patients with right and left-sided colon cancer. This was because there was a significant correlation between RANBP9 expression levels and tumor location. For patients in the right-sided colon cancer subgroup, low RANBP9 expression levels were not significantly correlated with DFS rates (P=0.171; Figure 2B). However, for patients in the left-sided colon cancer subgroup, low RANBP9 expression levels were significantly correlated with a worse DFS rate (P=0.037; Figure 2C). Among all the patients, 53 (23.2%) died during the follow-up period, of which 34 had low RANBP9 expression levels and 19 had high RANBP9 expression levels. Patients with low RANBP9 expression levels had a 5-year OS rate of 72.1% compared to 82.6% for patients with high RANBP9 expression levels (P=0.057; Figure 3).
Additional analysis of the clinicopathological parameters with regards to DFS and OS rates are summarized in Table 3. Cox’s proportional hazards model analysis revealed that RANBP9 expression levels (P=0.029, HR: 0.580, 95% CI: 0.356–0.946) and TNM stage (P=0.002, HR: 2.102, 95% CI: 1.302–3.392) had a significant influence on DFS rate. The univariate P-value of serum CEA was <0.1, and was included for multivariate analysis for OS rate. Multivariate analysis showed that RANBP9 expression levels (P=0.046, HR: 0.553, 95% CI: 0.309–0.989) and TNM staging (P=0.004, HR: 2.356, 95% CI: 1.315–4.222) significantly affected OS rates.
Table 3
| Variable | DFS | OS | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate P | Multivariate | Univariate P | Multivariate | ||||||
| HR | 95% CI | P | HR | 95% CI | P | ||||
| Gender: male vs. female | 0.904 | 0.427 | |||||||
| Age: <65 vs. ≥65 | 0.116 | 0.101 | |||||||
| Vascular invasion | 0.065 | 1.610 | 0.795–3.259 | 0.186 | 0.007 | 1.950 | 0.935–4.064 | 0.075 | |
| Perineural invasion | 0.867 | 0.385 | |||||||
| Serum CEA: normal vs. elevated | 0.208 | 0.049 | 1.735 | 0.937–3.214 | 0.080 | ||||
| Tumor location: right vs. left colon | 0.954 | 0.695 | |||||||
| Differentiation: well vs. poorly | 0.135 | 0.193 | |||||||
| T stage: T1, 2 vs. T3, 4 | 0.007 | 0.003 | |||||||
| N stage: N0 vs. N1, 2 | 0.011 | 0.006 | |||||||
| TNM: I, II vs. III, IV | 0.001 | 2.102 | 1.302–3.392 | 0.002 | 0.000 | 2.356 | 1.315–4.222 | 0.004 | |
| Post-surgical chemotherapy: no vs. yes | 0.164 | 0.399 | |||||||
| RANBP9: low vs. high expression | 0.017 | 0.580 | 0.356–0.946 | 0.029 | 0.061 | 0.553 | 0.309–0.989 | 0.046 | |
DFS, disease free survival; OS, overall survival; CEA, carcinoembryonic antigen; TNM, tumor node metastasis; RANBP9, RAN-binding protein 9.
Discussion
TNM is a staging method based on the human anatomy, however it occasionally fails to reflect the extent of tumor malignancy (21). Molecular staging, based on the identification of clustered genetic alterations and multi-marker phenotypes has gained a wider acceptance in recent years. It allows CRC to be stratified into diverse and discrete subtypes that evolved through independent evolutionary pathways (3,22). It has promising applicability in clinical studies and practice. In this study, we demonstrated the correlation between low RANBP9 expression levels with CRC clinical characteristics, including tumor location, vascular invasion, and normal serum CEA. Survival analysis demonstrated that low RANBP9 expression levels were associated with poor DFS and OS rates and was an independent prognostic factor for CRC survival as determined by multivariate Cox regression analysis. This finding is consistent with previous reports for osteosarcoma, lung, gastric and colorectal cancers, and demonstrated RANBP9 to be a protective factor (12-15,18). RANBP9 is involved in the mitochondrial apoptotic pathway during DNA damage-induced apoptosis. It inhibits the ERK pathway by interacting with the c-Raf complex, and complexes with TSSC3 to inhibit tumor metastases via the inhibition of SRC-mediated AKT signaling in osteosarcomas (12,23,24). This suggests that RANBP9 acts as a tumor suppressor. Consequently, high RANBP9 expression levels could be considered as a molecular marker for reduced tumor aggressiveness and may be a good CRC prognostic factor. However, Tessari et al. suggested a contradictory role for RANBP9 in tumorigenesis. In their study, RANBP9 acted as a positive mediator of DNA-damage response and had potential oncogenic activity (16,17). These differences may be cell-type specific, or dependent on tumor stage or other clinical characteristics.
Colorectal cancer has different clinicopathological features and genetic patterns in right and left-sided colon cancer and may be mainly because of their different embryonic origins. Several studies have reported that patients with right-sided colon cancer were older, more often female, had more advanced tumor stages, larger tumor size, and with often poorly differentiated tumors (25). Our study demonstrated that RANBP9 expression for right-sided colon cancer was lower compared to left-sided colon cancer, suggesting differences in biological characteristics. Since RANBP9 is thought to be a tumor suppressor, its lower expression in right-sided colon cancer compared to left-sided colon cancer in the present study is reasonable. Recent studies have shown that CRC prognosis is associated with the location of the primary tumor. It has been demonstrated that patients with left-sided colon cancer have a better prognosis compared to patients with right-sided colon cancer (26). Due to the limited sample size, we could not support these findings in our study using the Cox model. However, in subgroup analysis, RANBP9 expression was found to be beneficial for patient prognosis with left or the right-sided colon cancer, although the difference for the latter was not statistically significant.
Vascular invasion is critical for tumor cell dissemination and metastasis in a variety of cancer cells, and has been shown to be a poor prognostic factor for CRC (27). The American College of Pathologists classifies vascular invasion as a prognostic factor of category I (28). To invade tissues and organs, tumor cells must penetrate into the surrounding stroma and vessel wall, which consists of endothelial cells and extracellular matrix components. Integrin signaling pathway is essential for invasion and metastasis (29,30). It was found that RANBP9 was associated with LFA-1, an integrin receptor that has a role in cell adhesion and migration (31). Overexpression of RANBP9 has been demonstrated to inhibit cell adhesion. Knockdown of RANBP9 promoted adhesion, as well as spreading via integrin-dependent signaling (32). In gastric cancer cells, downregulation of RANBP9 decreased cell adhesion and increased cell motility (14). The role of RANBP9 in cell adhesion and migration may explain the relation between low RANBP9 expression levels and vascular invasion of CRC observed in the present study. However, vascular invasion was not an independent factor in survival analysis. This could be because of our small patient cohort size, and there were only 19 patients with vascular invasion.
Our study had several limitations. First, this was a retrospective study of post-surgical patients performed in a local medical institution. Hence, selection bias could not be avoided as the distribution of clinical characteristics of CRC patients in our study differs from the general epidemiological data of CRC to some extent (33); in addition, the association between RANBP9 and pre-surgical serum CEA levels in this study needs to be validated in multicenter institutions using larger patient cohorts. CEA is useful for post-surgical surveillance of CRC patients, however there have been some conflicting reports on pre-surgical CEA as a prognostic factor (34). Second, RANBP9 is a part of a protein complex that is larger than 670KD. It has several characteristics of a scaffold protein, including protein interactions, cytoskeleton binding, and several classical anchoring sites of signal transducers (35,36). Future studies are required to elucidate the mechanisms underlying its expression and its role in CRC.
Conclusions
Low RANBP9 expression levels were significantly correlated with tumor location, vascular invasion and poor survival in CRC patients. We identified low RANBP9 expression levels as a negative prognostic factor. Our findings may help advance the development of novel therapeutic strategies to improve CRC outcomes in patients.
Acknowledgments
The authors thank Dr. Weimin Ren for valuable assistance with immunohistochemistry.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.24). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Ethical approval was obtained from the Ethics Committee of Jinshan Hospital, Fudan University (Shanghai, China, No. 2011-02-01). Written informed consents for the use of tissue samples was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 2016;66:271-89. [Crossref] [PubMed]
- Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4.
- Belt EJ, van Stijn MF, Bril H, et al. Lymph node negative colorectal cancers with isolated tumor deposits should be classified and treated as stage III. Ann Surg Oncol 2010;17:3203-11. [Crossref] [PubMed]
- Barresi V, Reggiani Bonetti L, Ieni A, et al. Histologic prognostic markers in stage IIA colorectal cancer: a comparative study. Scand J Gastroenterol 2016;51:314-20. [Crossref] [PubMed]
- Carethers JM, Jung BH. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015;149:1177-90.e3. [Crossref] [PubMed]
- de Sousa E Melo F, Colak S, Buikhuisen J, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011;9:476-85. [Crossref] [PubMed]
- Summers MG, Smith CG, Maughan TS, et al. BRAF and NRAS Locus-Specific Variants Have Different Outcomes on Survival to Colorectal Cancer. Clin Cancer Res 2017;23:2742-9. [Crossref] [PubMed]
- Modest DP, Ricard I, Heinemann V, et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann Oncol 2016;27:1746-53. [Crossref] [PubMed]
- Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015;148:77-87.e2. [Crossref] [PubMed]
- Salemi LM, Maitland MER, McTavish CJ, et al. Cell signalling pathway regulation by RanBPM: molecular insights and disease implications. Open Biol 2017;7: [Crossref] [PubMed]
- Emberley ED, Gietz RD, Campbell JD, et al. RanBPM interacts with psoriasin in vitro and their expression correlates with specific clinical features in vivo in breast cancer. BMC Cancer 2002;2:28. [Crossref] [PubMed]
- Dai H, Lv YF, Yan GN, et al. RanBP9/TSSC3 complex cooperates to suppress anoikis resistance and metastasis via inhibiting Src-mediated Akt signaling in osteosarcoma. Cell Death Dis 2016;7:e2572. [Crossref] [PubMed]
- Zhu LL, Wang CH, Yang HP, et al. Expression of cartilage antitumor component RanBP9 in osteosarcoma. J Biol Regul Homeost Agents 2016;30:103-10. [PubMed]
- Shao S, Sun PH, Satherley LK, et al. Reduced RanBPM Expression Is Associated with Distant Metastasis in Gastric Cancer and Chemoresistance. Anticancer Res 2016;36:1295-303. [PubMed]
- Zhao Z, Cheng S, Zabkiewicz C, et al. Reduced Expression of RanBPM Is Associated with Poorer Survival from Lung Cancer and Increased Proliferation and Invasion of Lung Cancer Cells In Vitro. Anticancer Res 2017;37:4389-97. [PubMed]
- Palmieri D, Scarpa M, Tessari A, et al. Ran Binding Protein 9 (RanBP9) is a novel mediator of cellular DNA damage response in lung cancer cells. Oncotarget 2016;7:18371-83. [Crossref] [PubMed]
- Tessari A, Parbhoo K, Pawlikowski M, et al. RANBP9 affects cancer cells response to genotoxic stress and its overexpression is associated with worse response to platinum in NSCLC patients. Oncogene 2018;37:6463-76. [Crossref] [PubMed]
- Qin C, Zhang Q, Wu G. RANBP9 suppresses tumor proliferation in colorectal cancer. Oncol Lett 2019;17:4409-16. [PubMed]
- Jin GZ, Li Y, Cong WM, et al. iTRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J Proteome Res 2011;10:3418-28. [Crossref] [PubMed]
- Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 1987;8:138-40. [PubMed]
- Puppa G, Sonzogni A, Colombari R, et al. TNM staging system of colorectal carcinoma: a critical appraisal of challenging issues. Arch Pathol Lab Med 2010;134:837-52. [PubMed]
- Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017;17:79-92. [Crossref] [PubMed]
- Atabakhsh E, Bryce DM, Lefebvre KJ, et al. RanBPM has proapoptotic activities that regulate cell death pathways in response to DNA damage. Mol Cancer Res 2009;7:1962-72. [Crossref] [PubMed]
- Atabakhsh E, Schild-Poulter C. RanBPM is an inhibitor of ERK signaling. PLoS One 2012;7:e47803. [Crossref] [PubMed]
- Benedix F, Kube R, Meyer F, et al. Comparison of 17,641 patients with right- and left-sided colon cancer: differences in epidemiology, perioperative course, histology, and survival. Dis Colon Rectum 2010;53:57-64. [Crossref] [PubMed]
- Yahagi M, Okabayashi K, Hasegawa H, et al. The Worse Prognosis of Right-Sided Compared with Left-Sided Colon Cancers: a Systematic Review and Meta-analysis. J Gastrointest Surg 2016;20:648-55. [Crossref] [PubMed]
- Fujii T, Sutoh T, Morita H, et al. Vascular invasion, but not lymphatic invasion, of the primary tumor is a strong prognostic factor in patients with colorectal cancer. Anticancer Res 2014;34:3147-51. [PubMed]
- Compton CC, Fielding LP, Burgart LJ, et al. Prognostic factors in colorectal cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med 2000;124:979-94. [PubMed]
- Ganguly KK, Pal S, Moulik S, et al. Integrins and metastasis. Cell Adh Migr 2013;7:251-61. [Crossref] [PubMed]
- Fransvea E, Mazzocca A, Antonaci S, et al. Targeting transforming growth factor (TGF)-betaRI inhibits activation of beta1 integrin and blocks vascular invasion in hepatocellular carcinoma. Hepatology 2009;49:839-50. [Crossref] [PubMed]
- Denti S, Sirri A, Cheli A, et al. RanBPM is a phosphoprotein that associates with the plasma membrane and interacts with the integrin LFA-1. J Biol Chem 2004;279:13027-34. [Crossref] [PubMed]
- Woo JA, Roh SE, Lakshmana MK, et al. Pivotal role of RanBP9 in integrin-dependent focal adhesion signaling and assembly. FASEB J 2012;26:1672-81. [Crossref] [PubMed]
- Leung WK, Chen WQ, Gu L, et al. Distributions of colorectal cancer in two Chinese cities with contrasting colorectal cancer epidemiology. J Gastroenterol Hepatol 2015;30:1726-30. [Crossref] [PubMed]
- Gobbi PG, Valentino F, Berardi E, et al. New insights into the role of age and carcinoembryonic antigen in the prognosis of colorectal cancer. Br J Cancer 2008;98:328-34. [Crossref] [PubMed]
- Nishitani H, Hirose E, Uchimura Y, et al. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene 2001;272:25-33. [Crossref] [PubMed]
- Lakshmana MK, Chung JY, Wickramarachchi S, et al. A fragment of the scaffolding protein RanBP9 is increased in Alzheimer's disease brains and strongly potentiates amyloid-beta peptide generation. FASEB J 2010;24:119-27. [Crossref] [PubMed]


