
LncRNA MVIH knockdown inhibits the malignancy progression through downregulating miR-505 mediated HMGB1 and CCNE2 in acute myeloid leukemia
Introduction
Acute myeloid leukemia (AML) is a hematological malignancy characterized by malignant transformation of clonal neoplastic hematopoietic stem cells as well as the impaired normal hematopoiesis in the bone marrow and peripheral blood (1). Current therapies for AML commonly consist of intensive induction chemotherapy, stem cell transplantation and post-remission consolidation, which contributes to the complete remission of approximately 50–70% of adults with AML. However, only 20% of patients with AML enjoy long-term disease-free survival, and refractory or relapsed AML is the main cause of AML mortality (2,3). Existing evidence indicates that insight into genetic background of AML that foster the extensive understanding of AML pathology might lead to development of specific targeted therapy (4,5). Therefore, it is necessary to investigate the mechanism underlying the development and progression of AML to explore new therapeutic strategies.
Long non-coding RNAs (lncRNA) are defined as RNAs longer than 200 nucleotides, which participate in multiple stages of gene regulation including chromatin modification, regulation of protein modification, chromatin structure, and so on (6,7). Increasing number of researches reveal that lncRNAs also play important roles in diverse human diseases, especially cancers, and some lncRNAs, such as lnc-CCAT1, lnc-HOTAIR, are reported to exert functions through interacting with microRNA (miRNA) at the post-transcriptional level in AML (6,8,9). Recently, increasing studies indicate that lncRNA associated with microvascular invasion in hepatocellular carcinoma (lnc-MVIH) is upregulated and participates in the progression and development of several solid cancers, such as breast cancer, non-small cell lung cancer and hepatocellular carcinoma (10-13). However, little is known about its role in hematological malignancies. In addition, lnc-MVIH contains target sites for miR-505 as retrieved from miRanda database analysis (www.miranda.org), which is a tumor suppressor in several malignancies including chronic myeloid leukemia (14-16). Therefore, we hypothesized that lnc-MVIH might interact with miR-505 as well as miR-505 downstream target genes and regulate AML progression. In the present study, we investigated the regulatory role of lnc-MVIH in AML progression and the underlying mechanism, aiming to provide a novel insight of potential therapeutic targets in AML (17,18).
Methods
Cell sources and culture
Human AML cell lines AML-193, KG-1, HL-60 and OCI-AML2 were purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, German). AML-193 cells were cultured in 90% Iscove’s Modified Dulbecco’s Medium (Gibco, USA) and 10% fetal bovine serum (Gibco, USA); KG-1 and HL-60 cells were cultured in 90% Roswell Park Memorial Institute (RPMI) 1640 Medium (Gibco, USA) and 10% fetal bovine serum (Gibco, USA); OCI-AML2 were cultured in 90% Minimum Essential Medium (MEM) (Gibco, USA) and 10% fetal bovine serum (Gibco, USA).
Lnc-MVIH expression in AML cell lines and primary normal bone marrow mononuclear cells (BMMC)
Lnc-MVIH expression in AML cell lines AML-193, KG-1, HL-60 and OCI-AML2 was detected using real-time quantitative polymerase chain reaction (RT-qPCR). Besides, primary normal human BMMC was purchased from American type culture collection (Manassas, USA) and lnc-MVIH in BMMC was detected by RT-qPCR as control.
Lnc-MVIH expression in CD34+ AML cells and wild AML cell lines
CD34+ AML cells were separated from wild AML cell lines (AML-193, KG-1, HL-60, OCI-AML2) using Dynabeads CD34+ Positive Isolation Kit (Thermo, USA) according to the instructions of the manufacture, and lnc-MVIH expression in CD34+ AML cells and wild AML cells was detected using RT-qPCR.
Effect of lnc-MVIH knockdown on cell proliferation, apoptosis and potential target miR-505
Control shRNA [Control(−) group] and lnc-MVIH shRNA [lnc-MVIH(−) group] were constructed by Shanghai GenePharma Bio-Tech Company (Shanghai, China) using pGPH1 vector and transfected into KG-1 cells. Lnc-MVIH expression was detected at 24 h by RT-qPCR, cell proliferation was detected at 0, 24, 48, and 72 h by Counting Kit-8 (CCK-8) (Dojindo, Japan), cell apoptosis rate was detected at 24 h by Annexin V-FITC Apoptosis Detection Kit (BD, USA), potential target miR-505 was predicted using miRanda database analysis (www.miranda.org) and detected at 24 h by RT-qPCR, respectively.
Effect of miR-505 knockdown on attenuating lnc-MVIH knockdown functions
Lnc-MVIH shRNA [Lnc-MVIH(−) group], and lnc-MVIH shRNA plus miR-505 shRNA [Lnc-MVIH(−)&miR-505(−) group] were constructed by Shanghai GenePharma Bio-Tech Company (Shanghai, China) using pGPH1 vector and transfected into KG-1 cells. Lnc-MVIH and miR-505 expressions were detected at 24 h by RT-qPCR, miR-505 target genes including high mobility group box 1 (HMGB1) and cyclin E2 (CCNE2) were detected at 24 h by RT-qPCR and Western Blot, cell proliferation was detected at 0, 24, 48, and 72 h by CCK-8 (Dojindo, Japan), cell apoptosis rate was detected at 24 h by Annexin V-FITC Apoptosis Detection Kit (BD, USA).
RT-qPCR
Total RNA was extracted from cells using TRIzol™ Reagent (Invitrogen, USA) and then reversely transcribed to cDNA using RT-PCR Quick Master Mix (Toyobo, Japan). Following that, RT-qPCR was performed using SYBR® Green Realtime PCR Master Mix (Toyobo, Japan) to quantify lnc-MVIH, miR-505, HMGB1 and CCNE2 expressions. The result was calculated using 2−ΔΔCt method with GAPDH as an internal reference. The primers used in RT-qPCR were listed in Table 1.
Table 1
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| Lnc-MVIH | AATTTTGCACATCTGAACAGCC | TTCAAAATCCCACTACGCCCA |
| HMGB1 | TTCTTCCTCTTCTGCTCTGAGTATC | TTCATAAGGCTGCTTGTCATCTG |
| CCNE2 | GCTGCTGCCTTGTGCCATT | GTGCTCTTCGGTGGTGTCATAA |
| GAPDH | GACCACAGTCCATGCCATCAC | ACGCCTGCTTCACCACCTT |
| MiR-505 | ACACTCCAGCTGGGGGGAGCCAGGAAGTAT | TGTCGTGGAGTCGGCAATTC |
| U6 | CGCTTCGGCAGCACATATACTA | ATGGAACGCTTCACGAATTTGC |
Western blot
Total protein was extracted with RIPA Buffer (Sigma, USA). The protein concentration in each sample was then measured using the Pierce™ Rapid Gold BCA Protein Assay Kit (Thermo, USA). Twenty ug protein samples were added to NuPAGE™ 4–12% Bis-Tris Protein Gels (Thermo, USA) and transferred onto polyvinylidene fluoride membrane (Millipore, German). After blocking with BSA (Thermo, USA) for 2 h, the membranes were incubated with the primary antibodies overnight at 4 °C. Then, the membranes were incubated with the secondary antibody for 90 min at 37 °C. EasyBlot ECL kit (Sangon, China) was used to illuminized the bands and X-ray film (Kodak, USA) was used to visualize the result. The antibodies used in this study were summarized in Table 2.
Table 2
| Antibody | Company | Dilution |
|---|---|---|
| Primary antibody | ||
| Rabbit monoclonal to HMGB1 | Abcam (UK) | 1:2,000 |
| Rabbit monoclonal to Cyclin E2 | Abcam (UK) | 1:2,000 |
| Rabbit monoclonal to GAPDH | Abcam (UK) | 1:3,000 |
| Secondary antibody | ||
| Goat anti-rabbit IgG H&L (HRP) | Abcam (UK) | 1:5,000 |
Statistics
GraphPad 7.01 (GraphPad Int., USA) was used for statistical analysis and drawing images. Data were mainly presented as mean ± standard deviation, and compared by One-way ANOVA test followed by multiple comparisons test or t test. P<0.05 was considered as significant.
Results
The relative expression of lnc-MVIH in AML cell lines and primary normal BMMC
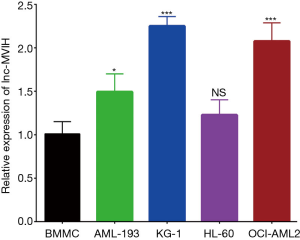
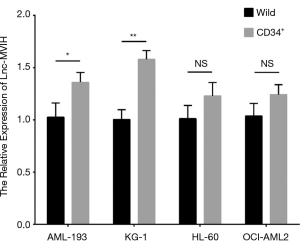
The relative expression of lnc-MVIH was elevated in AML-193 (P<0.05), KG-1 (P<0.001), OCI-AML2 (P<0.001) cells, but similar in HL-60 (P>0.05) cells compared with primary normal BMMC (Figure 1), indicating that lnc-MVIH was upregulated in AML cell lines.
The relative expression of lnc-MVIH in CD34+ and wild AML cells
The relative expression of lnc-MVIH was elevated in CD34+ AML-193 cells compared with wild AML-193 cells (P<0.05), and was also increased in CD34+ KG-1 cells compared with wild KG-1 cells (P<0.01), while similar between CD34+ HL-60 cells and wild HL-60 cells (P>0.05), between CD34+ OCI-AML2 cells and wild OCI-AML2 cells (P>0.05) (Figure S1).
The relative expression of lnc-MVIH after transfection
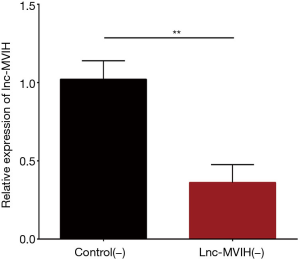
The expression of lnc-MVIH was decreased in lnc-MVIH(−) group compared with Control(−) group, which indicated the successful transfection of lnc-MVIH shRNA (P<0.01) (Figure 2).
Effect of lnc-MVIH knockdown on cell proliferation and apoptosis in KG-1 cells
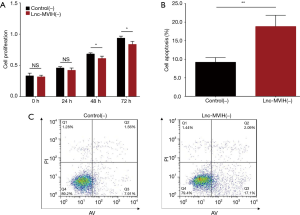
Cell proliferation was decreased in lnc-MVIH(−) group at 48 h (P<0.05) and 72 h (P<0.05), but unchanged at 0 h (P>0.05) and 24 h (P>0.05) compared with Control(−) group (Figure 3A). Cell apoptosis rate was increased in Lnc-MVIH(−) group compared with Control(−) group at 24 h (P<0.01) (Figure 3B,C). These data suggested that lnc-MVIH knockdown suppressed cell proliferation but promoted cell apoptosis in AML.
The relative expression of miR-505 after transfection in KG-1 cells
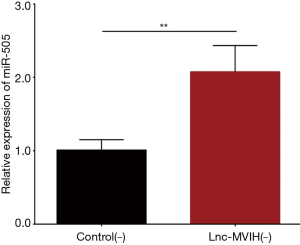
The relative expression of miR-505 was elevated in lnc-MVIH(−) group compared with Control(−) group after transfection (P<0.01), which indicated that lnc-MVIH knockdown promoted miR-505 expression in AML (Figure 4).
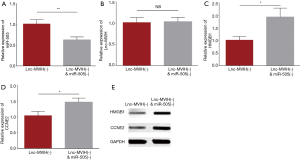
The effect of miR-505 knockdown on HMGB1 and CCNE2 expressions in lnc-MVIH knockdown treated KG-1 cells
The rescue experiments were conducted to explore the effect of miR-505 knockdown on HMGB1 and CCNE2 expressions in lnc-MVIH knockdown treated KG-1 cells. Firstly, we observed that the relative expression of miR-505 was decreased in lnc-MVIH(−) & miR-505(−) group compared with lnc-MVIH(−) group (P<0.01), which indicated the successful transfection of miR-505 shRNA (Figure 5A). Then the relative expression of lnc-MVIH was shown to be similar in both Lnc-MVIH(−) & miR-505(−) group and Lnc-MVIH(−) group (P>0.05), which suggested that miR-505 did not regulate lnc-MVIH (Figure 5B). Finally, HMGB1 and CCNE2, as the target genes of miR-505, were detected, which exhibited that the relative expressions of HMGB1 (P<0.05) (Figure 5C) and CCNE2 (P<0.05) (Figure 5D) were increased in lnc-MVIH(−) & miR-505(−) group compared with lnc-MVIH(−) group. And Western blot also visualized that HMGB1 and CCNE2 protein expression was increased in lnc-MVIH(−) & miR-505(−) group compared with lnc-MVIH(−) group (Figure 5E). These data suggested that miR-505 knockdown increased the expressions of HMGB1 and CCNE2 in lnc-MVIH knockdown treated AML cells.
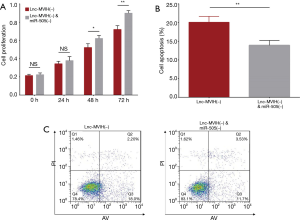
The effect of miR-505 knockdown on cell proliferation and cell apoptosis in lnc-MVIH knockdown treated KG-1 cells
Cell proliferation was increased in lnc-MVIH(−) & miR-505(−) group at 48 h (P<0.05) and 72 h (P<0.01), but unchanged at 0 h (P>0.05) and 24 h (P>0.05) compared with lnc-MVIH(−) group (Figure 6A). Cell apoptosis rate was decreased in lnc-MVIH(−) & miR-505(−) group compared with lnc-MVIH(−) group at 24 h (P<0.01) (Figure 6B,C). These data displayed that miR-505 knockdown compensated the effect of lnc-MVIH knockdown on cell proliferation and cell apoptosis in AML. Combining all the data above, we speculated that lnc-MVIH knockdown inhibited AML progression possibly via regulating miR-505 mediated HMGB1 and CCNE2 in AML.
Discussion
In the present study, we observed that lnc-MVIH was upregulated in AML cell lines compared with primary normal BMMC, and its knockdown suppressed cell proliferation but promoted cell apoptosis via regulating miR-505 mediated HMGB1 and CCNE2 in AML.
Lnc-MVIH is located in the intron of the RPS24 gene and is essential for transcription process of RPS24 (19). Existing studies demonstrate that it is upregulated and closely related with tumorigenesis and metastasis in some solid tumors (11-13,19). Lnc-MVIH is initially found to be upregulated in hepatocellular carcinoma and promotes tumor growth and metastasis by activating tumor-inducing angiogenesis (10). It is also indicated to be overexpressed in MDA-MB-231 breast cancer cell line compared to mammary epithelial cells and lnc-MVIH knockdown inhibits cell proliferation, increases cell apoptosis, elevates the G1–G0 phase cell proportion and decreases the percentage of cells in the S phase in MDA-MB-231 breast cancer cell line (13). In another study, lnc-MVIH is upregulated and its knockdown impaires cell proliferation as well as invasion in non-small cell lung cancer (12). Furthermore, the insufficiency of RPS24 gene, where lnc-MVIH is located on, is indicated to cause the cell cycle defects in hematological diseases such as diamond-Blackfan anemia (20). In addition, through the miRanda database analysis we found that lnc-MVIH contained several target sites of miR-505 which was shown to be biomarkers in chronic myeloid leukemia (20). Therefore, we hypothesized that lnc-MVIH might participate in the biological processes of AML. In our study, we observed that lnc-MVIH was overexpressed in AML cell lines compared with primary normal BMMC, and lnc-MVIH knockdown inhibited cell proliferation but promoted cell apoptosis in AML. The possible reasons might include that (I) lnc-MVIH might serve as the sponge for miRNA (such as miR-505), leading to the inhibitory effect on miRNA target genes (such as HMGB1 and CCNE2), contributing to the AML progression (16). (II) Lnc-MVIH knockdown might inactivate the oncogenic pathways such as WNT signaling pathway to inhibit the malignant progression (21).
Existing evidence exhibits that lnc-MVIH is involved in the regulation of miRNA target genes via serving as miRNA sponge in solid tumors (6). For example, lnc-MVIH knockdown inhibits the cell viability and promotes cell apoptosis through upregulating miR-199a and miR-199a targeted FZD7 in hepatocellular carcinoma (11). In addition, miRanda database analysis reveals that miR-505, which suppresses cell proliferation in hematological malignancies, is the potential target of lnc-MVIH (15,16,22). Then we detected the expression of miR-505 after transfecting lnc-MVIH shRNA into KG-1 cells and found that lnc-MVIH knockdown upregulated the expression of miR-505. Furthermore, HMGB1 and CCNE2 are potential target genes of miR-505, which are previously shown to be oncogenes in hematological malignancies (22,23). HMGB1, as a common oncogene in AML, triggers pro-inflammatory stimulus and activates the immune receptors, leading to the proliferation of AML cells, which is critical for the progression of AML (18,23,24). Besides, CCNE2 is often regarded as a key gene involved in cell cycle control, whose overexpression results in the proliferation advantage and malignancy progression in AML (17,25). Therefore, we conducted the rescue experiments to verify whether lnc-MVIH knockdown repressed cell activities via regulating miR-505 mediated HMGB1 and CCNE2, and found that miR-505 knockdown increased the expression of HMGB1 and CCNE2 in lnc-MVIH knockdown treated AML cells, meanwhile miR-505 knockdown compensated the effect of lnc-MVIH knockdown on cell proliferation and cell apoptosis in lnc-MVIH knockdown treated AML cells. Based on these data, we summarized that lnc-MVIH knockdown inhibited cell proliferation but promoted cell apoptosis via regulating miR-505 mediated HMGB1 and CCNE2. The possible reasons might include that: (I) lnc-MVIH knockdown impaired the expression of cell cycle regulatory proteins in the oncogenic pathways (such as Akt pathway, which CCNE2 participated in) via upregulation of miR-505, leading to the blocking effect of oncogenic pathway and inhibited cell proliferation but promoted cell apoptosis in AML (17). (II) Lnc-MVIH knockdown reduced the expression of HMGB1 and inhibited pro-inflammatory stimulus, which decreased proliferation and survival of AML cells (18).
In conclusion, lnc-MVIH knockdown inhibits cell proliferation but promotes cell apoptosis via regulating miR-505 mediated HMGB1 and CCNE2 in AML, which suggested that lnc-MVIH might serve as a potential therapeutic target for AML.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.12). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The institutional ethical approval and individual informed consent were waived due to the nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liao Q, Wang B, Li X, et al. miRNAs in acute myeloid leukemia. Oncotarget 2017;8:3666-82. [Crossref] [PubMed]
- Tallman MS, Gilliland DG, Rowe JM. Drug therapy for acute myeloid leukemia. Blood 2005;106:1154-63. [Crossref] [PubMed]
- Daver N, Cortes J. Molecular targeted therapy in acute myeloid leukemia. Hematology 2012;17:S59-62. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059-74. [Crossref] [PubMed]
- Schlenk RF. Post-remission therapy for acute myeloid leukemia. Haematologica 2014;99:1663-70. [Crossref] [PubMed]
- Wei S, Wang K. Long noncoding RNAs: pivotal regulators in acute myeloid leukemia. Exp Hematol Oncol 2016;5:30. [Crossref] [PubMed]
- Halley P, Kadakkuzha BM, Faghihi MA, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep 2014;6:222-30. [Crossref] [PubMed]
- Chen L, Wang W, Cao L, et al. Long Non-Coding RNA CCAT1 Acts as a Competing Endogenous RNA to Regulate Cell Growth and Differentiation in Acute Myeloid Leukemia. Mol Cells 2016;39:330-6. [Crossref] [PubMed]
- Xing CY, Hu XQ, Xie FY, et al. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett 2015;589:1981-7. [Crossref] [PubMed]
- Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients' poor recurrence-free survival after hepatectomy. Hepatology 2012;56:2231-41. [Crossref] [PubMed]
- Shi Y, Song Q, Yu S, et al. Microvascular invasion in hepatocellular carcinoma overexpression promotes cell proliferation and inhibits cell apoptosis of hepatocellular carcinoma via inhibiting miR-199a expression. Onco Targets Ther 2015;8:2303-10. [Crossref] [PubMed]
- Nie FQ, Zhu Q, Xu TP, et al. Long non-coding RNA MVIH indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumour Biol 2014;35:7587-94. [Crossref] [PubMed]
- Lei B, Xu SP, Liang XS, et al. Long non-coding RNA MVIH is associated with poor prognosis and malignant biological behavior in breast cancer. Tumour Biol 2016;37:5257-64. [Crossref] [PubMed]
- Tian L, Wang ZY, Hao J, et al. miR-505 acts as a tumor suppressor in gastric cancer progression through targeting HMGB1. J Cell Biochem 2018; [Epub ahead of print]. [PubMed]
- Lu L, Zhang D, Xu Y, et al. miR-505 enhances doxorubicin-induced cytotoxicity in hepatocellular carcinoma through repressing the Akt pathway by directly targeting HMGB1. Biomed. Pharmacother 2018;104:613-21. [Crossref] [PubMed]
- Ramachandran SS, Muiwo P, Ahmad HM, et al. miR-505-5p and miR-193b-3p: potential biomarkers of imatinib response in patients with chronic myeloid leukemia. Leuk Lymphoma 2017;58:1981-84. [Crossref] [PubMed]
- Payton M, Coats S. Cyclin E2, the cycle continues. Int J Biochem Cell Biol 2002;34:315-20. [Crossref] [PubMed]
- Yasinska IM, Goncalves Silva I, Sakhnevych SS, et al. High mobility group box 1 (HMGB1) acts as an "alarmin" to promote acute myeloid leukaemia progression. Oncoimmunology 2018;7:e1438109. [Crossref] [PubMed]
- He Y, Meng XM, Huang C, et al. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett 2014;344:20-27. [Crossref] [PubMed]
- Badhai J, Fröjmark AS, J Davey E, et al. Ribosomal protein S19 and S24 insufficiency cause distinct cell cycle defects in Diamond-Blackfan anemia. Biochim Biophys Acta 2009;1792:1036-42.
- Zhou HR, Fu HY, Wu DS, et al. Relationship between epigenetic changes in Wnt antagonists and acute leukemia. Oncol Rep 2017;37:2663-71. [Crossref] [PubMed]
- Lu L, Qiu C, Li D, et al. MicroRNA-505 suppresses proliferation and invasion in hepatoma cells by directly targeting high-mobility group box 1. Life Sci 2016;157:12-8. [Crossref] [PubMed]
- Yu Y, Xie M, He YL, et al. Role of high mobility group box 1 in adriamycin-induced apoptosis in leukemia K562 cells. Ai Zheng 2008;27:929-33. [PubMed]
- Tang D, Kang R, Zeh HJ 3rd, et al. High-mobility group box 1 and cancer. Biochim Biophys Acta 2010;1799:131-40.
- Qian Z, Joslin JM, Tennant TR, et al. Cytogenetic and genetic pathways in therapy-related acute myeloid leukemia. Chem Biol Interact 2010;184:50-7. [Crossref] [PubMed]


