
Analysis of volatile organic compounds released from SW480 colorectal cancer cells and the blood of tumor-bearing mice
Introduction
According to the International Agency for Research on Cancer (IRAC), approximately 1.36 million new colorectal cancer (CRC) cases were diagnosed worldwide in 2012, ranking third among malignant tumors (1). In 2017, 135,430 individuals in the United States were newly diagnosed with CRC, and 50,260 died from this disease (2). CRC is a high-risk cancer that is a serious threat to human health, as its morbidity and mortality are on the rise. The effective implementation of screening technology is a fundamental measure to reduce the incidence of CRC. Colonoscopy is considered the gold standard for screening CRC and advanced adenoma. Although this procedure has high sensitivity, it may cause bleeding, perforation and infection (3,4). Fecal immunochemical tests (FITs) are commonly used noninvasive screening methods. Although the sensitivity of FIT to CRC is approximately 70% (5-7), the sensitivity of advanced adenomas is not satisfactory (27–41%) (5,8-10). Therefore, an effective, noninvasive screening method is crucial for the prevention and treatment of CRC.
The analysis of volatile substances in metabolites is a noninvasive and promising new research strategy for identifying CRC and other cancers. Although volatile organic compound (VOC) analysis has not yet been applied in clinical practice, this method has been implemented in many studies (11-15). VOCs are produced by tumor cells, which exhibit an abnormal metabolism related to their unchecked growth. A second source of biomarkers related to CRC likely arises from the systemic effects of CRC and includes increased oxidative stress and increased catabolism. Studies have shown that various types of cancers, including lung cancer (14) and breast cancer (15), have specific VOC profiles that are different from those in the respective normal tissues. An impressive amount of data have confirmed the potential of some compounds to serve as a basis for a noninvasive, simple, inexpensive, easy-to-use diagnostic tool to assess cancer status.
Methods.
In vitro experiments
Cell culture
The SW480 cell line was obtained from the National Experimental Cell Resource Sharing Service Platform of the Chinese Academy of Medical Sciences. The SW480 CRC cell line was grown in RPMI 1640 culture medium supplemented with 10% fetal calf serum (FCS), 1% penicillin and 1% streptomycin. The cells were cultivated under standard conditions in a conventional incubator at 37 °C in a humidified atmosphere containing 5% CO2. When the cells reached 85–90% confluence, the culture medium was changed once, after which it was changed once every 2–3 days. To collect the VOCs produced by the cells, the supernatant in the culture flask was discarded, phosphate-buffered saline (PBS) free of calcium and magnesium ions was added to the flask to rinse the cells, and the PBS was discarded; the last two steps were performed twice. Trypsin was then added to the flask, which was incubated at 37 °C to release adherent cells. Immediately after digestion, 10 mL of phenol-red-free RPMI 1640 medium containing 10% fetal bovine serum was added to terminate the reaction. After thoroughly mixing the cell suspension, PBS was added to rinse the cells, and the suspension was centrifuged at 1,250 rpm/min for 5 min; these steps were performed twice. The PBS was discarded, and the resulting pellet was resuspended in 10 mL of phenol-red-free RPMI-1640 medium containing 10% fetal bovine serum.
Cell culture device
A device for culturing cells was developed (Figure 1). The cells were resuspended in 15 ml of phenol-red-free RPMI 1640 medium and seeded in a small Petri dish inside the glass culture apparatus. The long elbow (3) and short elbow (4) were placed in a small Petri dish (1), after which a beaker (5) was placed upside-down in the Petri dish (1) to keep the inside and outside of the beaker uniform. Then, to prevent cell contamination, a large Petri dish (6) was placed in the beaker, as shown in Figure 1, and the entire glass culture apparatus was placed in an incubator. After 24 hours of incubation, the cells were allowed to adhere, and the medium in the lower Petri dish was aspirated with a sterile Pasteur pipette and discarded. Afterwards, 80 mL of fresh medium (phenol-red-free RPMI 1640 medium supplemented with 10% FCS, 1% penicillin and 1% streptomycin) was added.
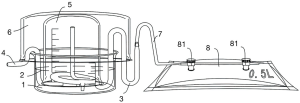
Addition of chemotherapeutic drugs to SW480 colorectal cancer cell culture media
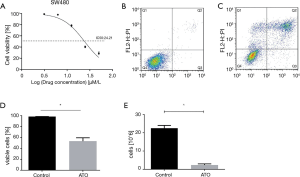
The IC50 values of arsenic trioxide (ATO) were measured in adherent cells to determine the optimal drug treatment concentration. Cell proliferation after ATO addition and adherent cell activity before and after ATO treatment were detected.
Sampling
Two milliliters of the culture supernatant were aspirated from the apparatus with a sterile syringe and immediately transferred to a vacuum-evaporated 20 mL glass vial (Supelco Inc., USA), upon which SPME-GC-MS was used to analyze the headspace. Before the medium was collected, all vials were thoroughly cleaned by flushing with nitrogen gas (purity of 99.9999%) to remove any residual contaminants, and the vials were then evacuated.
In vivo experiments
Inoculation of nude mice with SW480 cells
The Chinese Academy of Medical Sciences National Experimental Cell Resource Sharing Service Platform provided the SW480 human CRC cell line. Ten BALB/C nude/nude mice (female, 5–6 weeks old, 17–19 g, SPF grade) were purchased from Beijing Weitong Lihua Experimental Animal Technology Co., Ltd. The experiment was initiated 2 weeks after the animals arrived at our facility. The SW480 cells in logarithmic growth phase were prepared as a single cell suspension adjusted to 2×107/mL, and each nude mouse was injected in the rib and abdomen with 0.3 mL of the suspension via a disposable sterile syringe.
Preparation of transplanted tumors
The nude mice were inoculated with the SW480 cells and observed continuously for 4 weeks. After tumor formation, the mice were euthanized by cervical dislocation. After routine disinfection of the tumor site, any remaining necrotic tissue was removed. Tumor tissues that were light yellow in color or fish-like in appearance were placed in physiological saline and cut into approximately 2 mm pieces.
Sprague-Dawley rat model of colorectal cancer
Twenty healthy Sprague-Dawley rats (6 males, 250–300 g, SPF grade) were purchased from the Experimental Animal Center of the Second Affiliated Hospital of Harbin Medical University. The experiment was started 2 weeks after the animals arrived at our facility. A 2% sodium pentobarbital solution (50 mg/kg) was intraperitoneally injected, and the previously prepared tumor tissue was transplanted into the rat’s left armpit. Eight weeks after tumor inoculation, tumor resection was performed. The incision at the left armpit was used to find the site of tumor implantation, and the tumor tissue was isolated and completely resected.
Sampling
A total of 0.5 mL of blood was collected from the tail vein of the SD rats at the following times: before tumor inoculation (T0); 2 weeks (T2), 4 weeks (T4), 6 weeks (T6), and 8 weeks (T8) after inoculation; and 2 weeks (T10) and 4 weeks (T12) after tumor resection. The cells were immediately transferred to vacuum-evaporated 20 mL vials (Supelco Inc., USA) and subjected to SPME-GC-MS for headspace analysis. Before blood collection, all vials were thoroughly cleaned by flushing with nitrogen gas (purity of 99.9999%) to remove any residual contaminants and were then evacuated.
Solid-phase microextraction
Carboxy/polydimethylsiloxane SPME fiber (75 µm thick) was purchased from Supelco (Bellefonte, USA). The SPME fiber was inserted into a vial, and the sample was extracted at 40 °C for 40 minutes. Subsequently, the volatiles were thermally analyzed in a GC injector at 200 °C for 2 minutes.
GC/MS analysis
GC/MS analysis was performed on a Shimadzu GC-MS QP 2010 system (Shimadzu, Japan). The instrument was equipped with a DB-5MS unit with a length of 30 m, an ID of 0.25 µm and a thickness of 0.25 µm (Agilent Technologies, USA). Injection was performed in splitless mode. The injector temperature was 200 °C. The flow rate of the helium (99.999%) carrier gas was kept constant at 2 mL/min. The temperature of the column was maintained at 40 °C (1 min hold) to concentrate the hydrocarbons to the column head, increased by 5 °C min−1 to 200 °C (1 min hold), and then ramped by 15 °C min−1 to 230 °C. MS analysis was performed in full-scan mode using a scan range of 35–350 amu. The ion source was maintained at 230 °C using 70 eV of ionization energy for each measurement.
Extraction and pretreatment of raw GC-MS data
The raw data obtained by GC-MS were converted into the corresponding file format by the post-processing software that was included with the software. Then, the data were processed using the XCMS package in R software. The retention index of each peak was calculated from the ratio of the exact retention time of each peak and the existing retention time of the corresponding series of C4-C40 alkanes; then, the corresponding mass spectra of all the compounds were collected, and their corresponding retention time was determined. The accurate retention time index was compared with the mass spectrogram and retention time index of the National Institute of Standards and Technology (NIST) library 2.0 (2011) to identify the peaks.
Statistical analysis
Before conducting statistical analysis, we performed a total area normalization process on each sample and then output the normalized data to the SIMCA-P 11.5 platform for PCA and OPLSDA. To avoid overfitting, 100 iterations were used to perform the displacement test to verify the supervised model. To determine the significance of each metabolite, a nonparametric Kruskal-Wallis rank sum test was used. According to the predictor variable importance (VIP) calculated by the OPLSDA model and the non-parametric test P value, the potential metabolic biomarkers were chosen using threshold values of 1.0 and 0.05, respectively.
Results
The IC50 values of the drugs were determined after adding ATO to adherent cells (Figure 2A). The proliferation assay (Figure 2B) and activity detection (Figure 2C,D,E) were performed on the adherent cells. The IC50 value of the chemotherapeutic drug ATO in the SW480 cells was 24.21 µm/L; thus, the final ATO concentration in the SW480 cells in all subsequent experiments was 50 µm/L. The number of cells in the SW480 group was 23.30×106±1.57×106 cells after 24 hours of culture. After treatment with ATO (50 µm/L) for 24 hours, there were 2.34×106±0.67×106 cells.
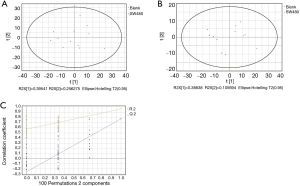
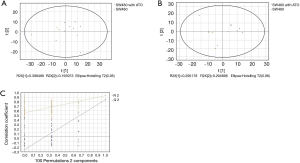
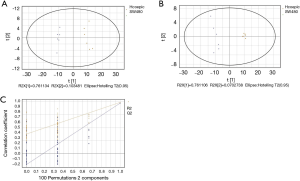
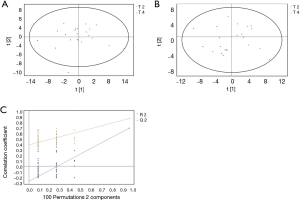
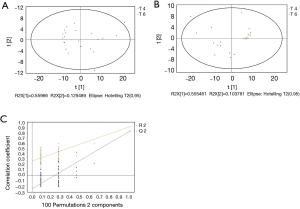
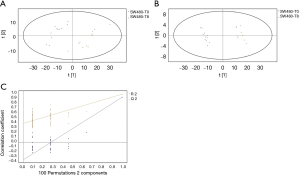
GC/MS was utilized to compare the metabolites in the medium between the blank group and the SW480 cell group (Figure 3), between the SW480 group and the SW480 with ATO group (Figure 4), between the HCoEpiC cell group (normal intestinal epithelium) and the SW480 cell group (Figure 5), between the T4 and T2 groups (Figure 6), between the T6 and T4 groups (Figure 7), between the T8 and T0 groups (Figure 8), between the T8 and T2 groups (Figure 9), between the T8 and T6 groups (Figure 10), between the T8 and T10 groups (Figure 11), between the T8 and T12 groups (Figure 12), and between the T10 group and T12 groups (Figure 13). The separation trend was detected from the PCA and OPLS-DA score plots. The tight clustering of samples in the OPLS-DA score plot demonstrates that our approach was effective.
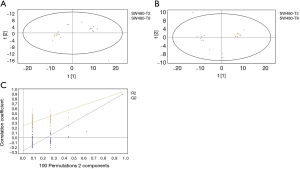
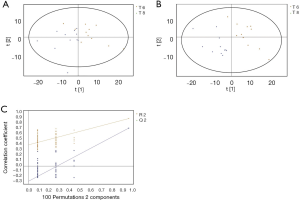
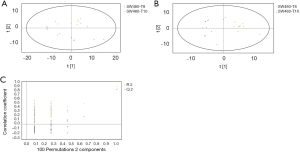
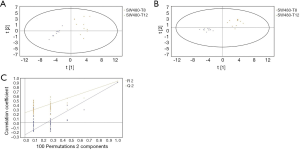
Potential markers
The potential biomarkers for CRC were determined by the VIP values obtained in the PLS-DA/OPLS-DA model and the P values in the nonreference test. To be identified as a potential biomarker, the VIP value must be greater than 1.0, and the P value must be less than 0.05. The structures of these compounds were identified by searching in the NIST library, and similarity was greater than 75%. The potential markers of the in vitro test groups are shown in Table 1, and those of the in vivo experimental group are shown in Table 2.
Table 1
| Potential biomarkers | P | FC | VIP | RT | Comparison point |
|---|---|---|---|---|---|
| Cyclohexanone | 0.00394 | 0.168471 | 1.56952 | 5.98059 | SW480(X2) vs. Blank(X1) |
| 1-Hexanol,2-ethyl- | 0.00394 | 0.236009 | 1.42799 | 9.75986 | SW480(X2) vs. Blank(X1) |
| 7-Octen-1-ol, 3,7-dimethyl-,(s)- | 0.00394 | 0.210518 | 1.67467 | 11.05258 | SW480(X2) vs. Blank(X1) |
| (+)-2-Bronanone | 0.02497 | 0.175680 | 1.12611 | 13.23279 | SW480(X2) vs. Blank(X1) |
| Levomenthol | 0.00394 | 0.236097 | 1.39101 | 14.14689 | SW480(X2) vs. Blank(X1) |
| Cyclohexanol,5-methyl-2-(1-methylethyl)-, [1R-(1.alpha.,2.beta.,5.alpha.)]- | 0.00394 | 0.236097 | 1.39101 | 14.14689 | SW480(X2) vs. Blank(X1) |
| Propanoic acid, 2-methyl-, 2,2-dimethyl-1-(2-hydroxy-1-methylethyl)propyl ester | 0.00395 | −0.444730 | 1.69848 | 18.83550 | SW480(X2) vs. Blank(X1) |
| Silane, cyclohexyldimethoxymethyl- | 0.01041 | −0.187628 | 1.28663 | 13.50091 | SW480(X2) vs. Blank(X1) |
| Propanoic acid, 2-methyl-,3-hydroxy-2,4,4-trimethylpentyl ester | 0.00394 | −0.157310 | 1.40656 | 19.48938 | SW480(X2) vs. Blank(X1) |
| 2,5-Cyclohexadiene-1,4-dione, 2,6-bis(1,1-dimethylethyl)- | 0.00394 | −0.484064 | 1.6757 | 21.75504 | SW480(X2) vs. Blank(X1) |
| Pentanoic acid, 2,2,4-trimethyl-3-carboxyisopropyl, isobutyl ester | 0.00394 | −0.280585 | 1.65834 | 24.85678 | SW480(X2) vs. Blank(X1) |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, (2R-cis)- | 0.00395 | 0.662331 | 1.98593 | 13.51363 | SW480(X2) vs. ATO(X1) |
| p-Trimethylsilyloxyphenyl-(trimethylsilyloxy)trimethylsilylacrylate | 0.01631 | 0.206226 | 1.36752 | 12.94817 | SW480(X2) vs. ATO(X1) |
| Camphor | 0.02497 | 0.237656 | 1.28867 | 13.23732 | SW480(X2) vs. ATO(X1) |
| Butylated Hydroxytoluene | 0.03737 | 0.252073 | 1.34922 | 22.83002 | SW480(X2) vs. ATO(X1) |
| Cyclohexanol | 0.00394 | 0.811823 | 1.10983 | 5.85873 | SW480(X2) vs. Normal(X1) |
| 1-Hexanol, 2-ethyl- | 0.00394 | 1.383279 | 1.14552 | 9.79397 | SW480(X2) vs. Normal(X1) |
| Cyclohexanone, 5-methyl-2-(1-methylethyl)-, (2R-cis)- | 0.00394 | 1.009309 | 1.13705 | 13.52999 | SW480(X2) vs. Normal(X1) |
| Butylated Hydroxytoluene | 0.00388 | 1.903207 | 1.1067 | 22.85199 | SW480(X2) vs. Normal(X1) |
RT, retention time; VIP, variable importance in projection; FC, fold change, defined as: FC = log10 (X2/X1). A positive FC indicates that the concentrations of certain metabolites in the X2 group are relatively higher than those in the X1 group, whereas a negative FC value indicates that the concentrations of certain metabolites in the X2 group are lower than those in the X1 group.
Table 2
| Potential biomarkers | P | FC | VIP | RT | Comparison point |
|---|---|---|---|---|---|
| 1,3-Dioxane-4,6-dione, 2,2-dimethyl- | 0.00525 | 0.241042 | 1.25439 | 1.87855 | T4(X2) vs. T2(X1) |
| Silanediol, dimethyl- | 0.00144 | 0.495456 | 1.2059 | 2.89706 | T4(X2) vs. T2(X1) |
| Acetone | 0.00007 | −0.542360 | 1.93417 | 1.76784 | T4(X2) vs. T2(X1) |
| Oxime-, methoxy-phenyl-_ | 0.00091 | −0.197124 | 1.41167 | 6.40241 | T4(X2) vs. T2(X1) |
| 2-Octen-1-ol, (E)- | 0.00044 | −0.525596 | 1.539 | 8.20278 | T4(X2) vs. T2(X1) |
| Cyclotetrasiloxane, octamethyl- | 0.02782 | −0.054072 | 1.07055 | 8.29604 | T4(X2) vs. T2(X1) |
| l-Alanine ethylamide | 0.00525 | −0.137827 | 1.60883 | 33.76522 | T4(X2) vs. T2(X1) |
| Acetone | 0.00007 | 0.704458 | 1.68682 | 2.04834 | T6(X2) vs. T4(X1) |
| Pentadecane, 2,6,10,14-tetramethyl- | 0.00012 | 1.246748 | 1.09185 | 27.56510 | T6(X2) vs. T4(X1) |
| Tritetracontane | 0.00007 | 1.066486 | 1.09165 | 29.79724 | T6(X2) vs. T4(X1) |
| Silanediol, dimethyl- | 0.00020 | −0.945767 | 1.32604 | 3.09105 | T6(X2) vs. T4(X1) |
| 8-Hydroxy-2-octanone | 0.00115 | −0.484751 | 1.10374 | 6.38218 | T6(X2) vs. T4(X1) |
| 2-Heptanone | 0.00144 | 0.939597 | 1.03953 | 5.90268 | T8(X2) vs. T0(X1) |
| 3-Heptanone | 0.00115 | 0.918446 | 1.35697 | 9.57596 | T8(X2) vs. T0(X1) |
| Ether, 6-methylheptyl vinyl | 0.00115 | 0.918446 | 1.35697 | 9.59138 | T8(X2) vs. T0(X1) |
| Propanoic acid, 2-methyl-, 2,2-dimethyl-1-(2-hydroxy-1-methylethyl)propyl ester | 0.00091 | 0.646958 | 1.26083 | 18.85799 | T8(X2) vs. T0(X1) |
| Cyclotrisiloxane, hexamethyl- | 0.00007 | −0.871712 | 1.75269 | 4.21080 | T8(X2) vs. T0(X1) |
| 2-Pentanone | 0.00009 | −0.475081 | 1.60069 | 2.91339 | T8(X2) vs. T0(X1) |
| Cyclotetrasiloxane, octamethyl- | 0.00007 | −0.557822 | 1.83156 | 8.47760 | T8(X2) vs. T0(X1) |
| 1-Decanol, 2-hexyl- | 0.00281 | −0.689802 | 1.26712 | 26.27369 | T8(X2) vs. T0(X1) |
| p-Trimethylsilyloxyphenyl-(trimethylsilyloxy)trimethylsilylacrylate | 0.00007 | −0.746933 | 1.92595 | 12.96518 | T8(X2) vs. T0(X1) |
| Camphor | 0.00775 | −0.494386 | 1.2059 | 13.25408 | T8(X2) vs. T0(X1) |
| Acetone | 0.00007 | 0.400951 | 1.4991 | 2.02597 | T8(X2) vs. T2(X1) |
| 1-Hexanol, 2-ethyl- | 0.01974 | 0.394020 | 1.0091 | 9.83215 | T8(X2) vs. T2(X1) |
| Pentanoic acid, 2,2,4-trimethyl-3-hydroxy-, isobutyl ester | 0.00012 | 1.044741 | 1.1809 | 18.85967 | T8(X2) vs. T2(X1) |
| Butylated Hydroxytoluene | 0.00020 | 1.075230 | 1.1866 | 22.83927 | T8(X2) vs. T2(X1) |
| Cyclotrisiloxane, hexamethyl- | 0.00007 | −0.574909 | 1.39181 | 4.21830 | T8(X2) vs. T2(X1) |
| Cyclotetrasiloxane, octamethyl- | 0.00007 | −0.574119 | 1.41831 | 8.47704 | T8(X2) vs. T2(X1) |
| p-Trimethylsilyloxyphenyl-(trimethylsilyloxy)trimethylsilylacrylate | 0.00115 | −0.347959 | 1.16289 | 12.97072 | T8(X2) vs. T2(X1) |
| Acetone | 0.00007 | 0.612645 | 1.60045 | 2.04646 | T8(X2) vs. T6(X1) |
| Oxime-, methoxy-phenyl-_ | 0.00072 | 0.870624 | 1.34098 | 6.35366 | T8(X2) vs. T6(X1) |
| Arsenous acid, tris(trimethylsilyl) ester | 0.00007 | 0.696353 | 1.4879 | 8.13797 | T8(X2) vs. T6(X1) |
| Propanoic acid, 2-methyl-, 2,2-dimethyl-1-(2-hydroxy-1-methylethyl)propyl ester | 0.00007 | 0.561052 | 1.21175 | 18.86690 | T8(X2) vs. T6(X1) |
| l-Alanine ethylamide, (S)- | 0.00281 | 0.161825 | 1.2691 | 33.98259 | T8(X2) vs. T6(X1) |
| Silanediol, dimethyl- | 0.00091 | −2.272951 | 1.44175 | 2.34641 | T8(X2) vs. T6(X1) |
| Cyclotrisiloxane, hexamethyl- | 0.00020 | −0.387547 | 1.19322 | 4.18962 | T8(X2) vs. T6(X1) |
| 1-Octen-3-ol | 0.00115 | −0.307459 | 1.10375 | 8.39946 | T8(X2) vs. T6(X1) |
| Amyl vinyl carbinol | 0.00115 | −0.307459 | 1.10375 | 8.39946 | T8(X2) vs. T6(X1) |
| Cyclotetrasiloxane, octamethyl- | 0.00012 | −0.224971 | 1.43139 | 8.48799 | T8(X2) vs. T6(X1) |
| Hentriacontane | 0.00007 | −1.098505 | 1.23849 | 26.29285 | T8(X2) vs. T6(X1) |
| Hexadecane, 2,6,10,14-tetramethyl- | 0.00007 | −1.095723 | 1.17827 | 27.53987 | T8(X2) vs. T6(X1) |
| Pentadecane, 2,6,10,14-tetramethyl- | 0.00007 | −0.891132 | 1.20079 | 27.55948 | T8(X2) vs. T6(X1) |
| Tritetracontane | 0.00007 | −1.114259 | 1.17723 | 29.78725 | T8(X2) vs. T6(X1) |
| Acetone | 0.00006 | 0.776799 | 1.98977 | 2.05 | T8(X2) vs. T10(X1) |
| Arsenous acid, tris(trimethylsilyl) ester | 0.00007 | 0.518305 | 1.8232 | 8.13985 | T8(X2) vs. T10(X1) |
| Cyclotetrasiloxane, octamethyl- | 0.04520 | 0.178516 | 1.07588 | 8.48989 | T8(X2) vs. T10(X1) |
| 1-Octanol, 2-butyl- | 0.00417 | 0.353719 | 1.28211 | 9.81666 | T8(X2) vs. T10(X1) |
| Propanoic acid, 2-methyl-, 2,2-dimethyl-1-(2-hydroxy-1-methylethyl)propyl ester | 0.01123 | 0.348720 | 1.02944 | 18.86792 | T8(X2) vs. T10(X1) |
| Pentanoic acid, 2,2,4-trimethyl-3-carboxyisopropyl, isobutyl ester | 0.00513 | 0.550914 | 1.05131 | 24.89157 | T8(X2) vs. T10(X1) |
| l-Alanine ethylamide, (S)- | 0.00115 | 0.190858 | 1.59258 | 33.99298 | T8(X2) vs. T10(X1) |
| Toluene | 0.04520 | −2.139063 | 1.26771 | 3.33288 | T8(X2) vs. T10(X1) |
| Acetone | 0.00006 | 0.627783 | 1.40172 | 2.04766 | T8(X2) vs. T12(X1) |
| Ether, 6-methylheptyl vinyl | 0.00006 | 0.733642 | 1.18843 | 9.58711 | T8(X2) vs. T12(X1) |
| Arsenous acid, tris(trimethylsilyl) ester | 0.00007 | 0.545805 | 1.30776 | 8.13499 | T8(X2) vs. T12(X1) |
| Cyclotetrasiloxane, octamethyl- | 0.00007 | 0.155689 | 1.12794 | 8.48488 | T8(X2) vs. T12(X1) |
| 1-Hexanol, 2-ethyl- | 0.00006 | 0.810063 | 1.31477 | 9.81297 | T8(X2) vs. T12(X1) |
| Butylated Hydroxytoluene | 0.00006 | 0.673286 | 1.08778 | 22.84221 | T8(X2) vs. T12(X1) |
| 2-Heptanone | 0.00115 | 1.837936 | 1.14065 | 5.91052 | T10(X2) vs. T12(X1) |
| Butylated Hydroxytoluene | 0.00043 | 2.703213 | 4.2667 | 22.83680 | T10(X2) vs. T12(X1) |
RT, retention time; VIP, variable importance in projection; FC, fold change, defined as: FC = log10 (X2/X1). A positive FC indicates that the concentrations of certain metabolites in the X2 group are relatively higher than those in the X1 group, whereas a negative FC value indicates that the concentrations of certain metabolites in the X2 group are lower than those in the X1 group.
Discussion
Combining the in vivo and in vitro groups of colorectal cancer, we obtained a variety of potential markers, which can be divided into hydrocarbons, alcohols, ketones, esters, and aromatic compounds according to their chemical properties. At present, the potential metabolic pathways of these markers are not yet clear, and the metabolic sources of the detected substances are still speculative; however, a metabolic pathway [e.g., oxidative stress (16)] or enzyme [e.g., cytochrome p-450 enzyme (17)] may explain this process of change. Oxidative stress in the body is related to the formation and inactivation of reactive oxygen species (ROS) and free radicals. During the lipid peroxidation of fatty acid components of the ROS-initiated membrane, hydrocarbons (such as ethane, pentane and aldehydes) are formed. These hydrocarbons are considered markers of oxidative stress. ROS exist in different forms, such as hydroxyls (HO), superoxide (O2), peroxyls (ROO), alkoxyls (RO), and nitric oxides (NO). ROS may also originate from exogenous sources such as cigarette smoke, pollution, and radiation. Once accumulated in the tissue, ROS attack different molecules in the body, such as polyunsaturated fatty acids (PUFAs) and proteins. During oxidative stress, ROS and free radicals are excreted from the mitochondria of cells, producing volatile alkanes that are released during respiration (18). Protein oxidation may produce fewer metabolites, which are less soluble in the blood and are therefore excreted through exhalation. Moreover, important risk factors for cancer development are related to the induction of cytochrome p-450 enzymes (CYP450, a group of oxidases); in particular, cytochrome p-450 enzymes that catalyze the oxidation of organic chemicals can be upregulated by ROS molecules in human tissues (19). This enzyme family, which includes aromatase, an enzyme that synthesizes estrogen, has been shown to be overexpressed in human breast cancer tissues (20).
In our experiment, the concentration of butylated hydroxytoluene in the SW480 group was higher than those in the blank group and ATO group. The in vivo experiments showed that the concentration of butylated hydroxytoluene was higher at T8 than at T2, at T8 than at T12, and at T10 than at T12. This substance was shown to be highly concentrated in cancer cells, and its concentration decreased after the addition of ATO. After tumor resection, the concentration of butylated hydroxytoluene gradually decreased with time (higher at T10 than at T12). Butylated hydroxytoluene is an antioxidant with significant free radical scavenging capacity. ROS and free radicals play an important physiological role in the oxidation process, but they may concurrently exert toxic effects throughout the body. Free radicals not only cause premature aging and the appearance of wrinkles but also are the main cause of cancer and other chronic diseases (21). Our body has adapted by developing defense systems to reduce the damage caused by ROS (22). Antioxidants are the main defense mechanism for scavenging free radicals in the human body and can prevent damage caused by free radicals (23). Therefore, we speculate that butylated hydroxytoluene, an antioxidant, plays an important role in tumor growth and necrosis. The concentration of butylated hydroxytoluene may increase during tumor growth because the body initiates an antioxidant response to combat the metabolic changes caused by cancer. The concentration of antioxidants may decrease after treatment with chemotherapeutic drugs or tumor tissue resection because of the necrosis of tumor cells, at which point the body stops or slows the antioxidant response. As a result, the concentration of butylated hydroxytoluene and its antioxidant effects are reduced.
We found that acetone exhibited the following concentration profile in vivo: T2>T4, T6>T4, T8>T2, T8>T4, T8>T6, T8>T10, and T8>T12. The concentration of acetone gradually decreased at T2-T4 and gradually increased at T4-T8, but this activity was not exhibited in the in vitro experimental group. In vivo, the liver produces large amounts of acetoacetate and beta-hydroxybutyrate. Acetoacetic acid undergoes spontaneous decarboxylation to acetone (18). Due to the high vapor pressure of acetone, it is generated in smaller amounts than those of other ketone bodies and can be exhaled through breathing. Ketone bodies are also derived from amino acid metabolism. Under physiologically normal conditions, the rate of tissue protein catabolism is relatively constant throughout the day. However, ketone bodies and acetone generally form during the final stage of the disease, which can explain the inconsistency among different studies (24,25). The significant increase in the concentration of acetone in late inoculated tumors may be due to the accelerated rate of protein degradation and the acceleration of amino acid metabolism resulting in a large number of ketone bodies. Thus, acetone may be a potential marker of advanced cancer. The decrease in acetone concentration at T2-T4 may be due to other factors, such as eating and physical activity. We found that in the in vitro experiments, other ketones were expressed more highly in the SW480 group than in the blank, normal intestinal epithelial and chemotherapy groups. These compounds share a common molecule: cyclohexanone. Researchers have shown that cyclohexanone is elevated in the headspace of urine in patients with kidney cancer (26). Westhoff et al. have shown that healthy individuals and COPD patients can be distinguished by the concentration of cyclohexanone upon exhalation (27). Therefore, whether cyclohexanone is a viable marker remains to be discussed.
The in vivo experiments revealed a variety of alkane compounds, such as silane, cyclohexyldimethoxymethyl-; cyclotrisiloxane, hexamethyl-; cyclotetrasiloxane, octamethyl-; hexadecane, 2,6,10,14-tetramethyl-; pentadecane, and 2,6,10,14-tetramethyl-. Many studies have shown that alkanes can be used as potential markers for lung cancer (28), breast cancer (29), and head and neck cancer (30). The key mechanism involved in the production of hydrocarbons in the body is oxidative stress. Alkanes are mainly produced by the peroxidation of polyunsaturated fatty acids. Lipid peroxidation is involved in tissue damage in vivo and may contribute to the development of cancer, inflammatory diseases, atherosclerosis, and aging. The body attempts to control and reduce lipid peroxidation by producing antioxidants (18). Hydrocarbons such as ethane and pentane are the final products of the lipid peroxidation process. The levels of ethane and pentane in exhaled breath have been widely used as noninvasive indicators of lipid peroxidation in vivo (31).
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2019.10.21). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Ethics Committee at the First Affiliated Hospital of Harbin Medical University (No. 2017005) in compliance with the institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- International Agency For Research on Cancer. Colorectal cancer estimated incidence, mortality and prevalence worldwide in 2012. Available online: https://globocan.iarc.fr/Pages/fact_sheets_cancer.asps.
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal Cancer Statistics. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Arora G, Mannalithara A, Singh G, et al. Risk of perforation from a colonoscopy in adults: a large population-based study. Gastrointest Endosc 2009;69:654-64. [Crossref] [PubMed]
- Levin TR, Zhao W, Conell C, et al. Complications of colonoscopy in an integrated health care delivery system. Ann Intern Med 2006;145:880-6. [Crossref] [PubMed]
- Terhaar sive Droste JS, Oort FA, van der Hulst RW, et al. Higher fecal immunochemical test cutoff levels: lower positivity rates but still acceptable detection rates for early-stage colorectal cancers. Cancer Epidemiol Biomarkers Prev 2011;20:272-80.
- Oort FA, Terhaar Sive Droste JS, Van Der Hulst RW, et al. Colonoscopy-controlled intra-individual comparisons to screen relevant neoplasia: faecal immunochemical test vs. guaiac-based faecal occult blood test. Aliment Pharmacol Ther 2010;31:432-9. [Crossref] [PubMed]
- de Wijkerslooth TR, Stoop EM, Bossuyt PM, et al. Immunochemical fecal occult blood testing is equally sensitive for proximal and distal advanced neoplasia. Am J Gastroenterol 2012;107:1570-8. [Crossref] [PubMed]
- Levi Z, Rozen P, Hazazi R, et al. A quantitative immunochemical fecal occult blood test for colorectal neoplasia. Ann Intern Med 2007;146:244-55. [Crossref] [PubMed]
- Rozen P, Levi Z, Hazazi R, et al. Identification of colorectal adenomas by a quantitative immunochemical faecal occult blood screening test depends on adenoma characteristics, development threshold used and number of tests performed. Aliment Pharmacol Ther 2009;29:906-17. [Crossref] [PubMed]
- Haug U, Kuntz KM, Knudsen AB, et al. Sensitivity of immunochemical faecal occult blood testing for detecting left- vs right-sided colorectal neoplasia. Br J Cancer 2011;104:1779-85. [Crossref] [PubMed]
- Tong H, Wang Y, Li Y, et al. Volatile organic metabolites identify patients with gastric carcinoma, gastric ulcer, or gastritis and control patients. Cancer Cell Int 2017;17:108. [Crossref] [PubMed]
- Wang D, Wang C, Pi X, et al. Urinary volatile organic compounds as potential biomarkers for renal cell carcinoma. Biomed Rep 2016;5:68-72. [Crossref] [PubMed]
- Guo L, Wang C, Chi C, et al. Exhaled breath volatile biomarker analysis for thyroid cancer. Transl Res 2015;166:188-95. [Crossref] [PubMed]
- Wang C, Dong R, Wang X, et al. Exhaled volatile organic compounds as lung cancer biomarkers during one-lung ventilation. Sci Rep 2014;4:7312. [Crossref] [PubMed]
- Wang C, Sun B, Guo L, et al. Volatile organic metabolites identify patients with breast cancer, cyclomastopathy, and mammary gland fibroma. Sci Rep 2014;4:5383. [Crossref] [PubMed]
- Ambrosone CB. Oxidants and antioxidants in breast cancer. Antioxid Redox Signal 2000;2:903-17. [Crossref] [PubMed]
- Murray GI. The role of cytochrome P450 in tumour development and progression and its potential in therapy. J Pathol 2000;192:419-26. [Crossref] [PubMed]
- Hakim M, Broza YY, Barash O, et al. Volatile organic compounds of lung cancer and possible biochemical pathways. Chem Rev 2012;112:5949-66. [Crossref] [PubMed]
- Watanabe M. Polymorphic CYP genes and disease predisposition--what have the studies shown so far? Toxicol Lett 1998;102-103:167-71. [Crossref] [PubMed]
- Chen S. Aromatase and breast cancer. Front Biosci 1998;3:d922-33. [Crossref] [PubMed]
- Valko M, Leibfritz D, Moncol J, et al. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 2007;39:44-84. [Crossref] [PubMed]
- Maltepe E, Saugstad OD. Oxygen in health and disease: regulation of oxygen homeostasis--clinical implications. Pediatr Res 2009;65:261-8. [Crossref] [PubMed]
- Pham-Huy LA, He H, Pham-Huy C. Free radicals, antioxidants in disease and health. Int J Biomed Sci 2008;4:89-96. [PubMed]
- Bajtarevic A, Ager C, Pienz M, et al. Noninvasive detection of lung cancer by analysis of exhaled breath. BMC Cancer 2009;9:348. [Crossref] [PubMed]
- Kischkel S, Miekisch W, Sawacki A, et al. Breath biomarkers for lung cancer detection and assessment of smoking related effects--confounding variables, influence of normalization and statistical algorithms. Clin Chim Acta 2010;411:1637-44. [Crossref] [PubMed]
- Thompson-Torgerson CS, Champion HC, Santhanam L, et al. Cyclohexanone contamination from extracorporeal circuits impairs cardiovascular function. Am J Physiol Heart Circ Physiol 2009;296:H1926-32. [Crossref] [PubMed]
- Westhoff M, Litterst P, Maddula S, et al. Differentiation of chronic obstructive pulmonary disease (COPD) including lung cancer from healthy control group by breath analysis using ion mobility spectrometry. Int J Ion Mobil Spectrom 2010;13:131-9. [Crossref]
- Poli D, Carbognani P, Corradi M, et al. Exhaled volatile organic compounds in patients with non-small cell lung cancer: cross sectional and nested short-term follow-up study. Respir Res 2005;6:71. [Crossref] [PubMed]
- Phillips M, Cataneo RN, Saunders C, et al. Volatile biomarkers in the breath of women with breast cancer. J Breath Res 2010;4:026003. [Crossref] [PubMed]
- Hakim M, Billan S, Tisch U, et al. Diagnosis of head-and-neck cancer from exhaled breath. Br J Cancer 2011;104:1649-55. [Crossref] [PubMed]
- Terelius Y, Ingelman-Sundberg M. Metabolism of n-pentane by ethanol-inducible cytochrome P-450 in liver microsomes and reconstituted membranes. Eur J Biochem 1986;161:303-8. [Crossref] [PubMed]


