
CRISPR-Cas genome editing tool: a narrow lane of cancer therapeutics with potential blockades
About CRISPR-Cas9 genome editing technology
From precellular evolution to cellular evolution, the contribution of RNA world theory is well appreciated. In recent decades, the discovery of a key components in adaptive immune systems of bacteria has revolutionized cellular and molecular biology (1,2). A discovery of clustered regularly interspaced short palindromic repeats (CRISPR)-associated nucleases (Cas) system in bacterial defense system that employs both and RNA and proteins to achieve precision and efficient genome engineering (2-9). In fact, such discovery strengthens the importance of RNA in cellular evolution and adaptations. To remove ambiguity among reported CRISPR-Cas systems, three major types are denoted with signature genes such as Cas3 in type I systems, Cas9 in type II, and Cas10 in type III systems (9-15). The common feature among these CRISPR-Cas systems is to be seen as one of RNA based adaptable defense system in bacteria. In essence, CRISPR-Cas9 system is elegantly shown to employ two RNAs as crRNA that complex with a tracrRNA to achieve the targeting of nuclease Cas9 to specific DNA sequences. Actually, fusion of tracrRNA and crRNA is designated as single guide RNA (sgRNA) that directs CRISPR-Cas9 nuclease to position on target DNA sequence (12-15). In other way, Cas9 nuclease in complex with crRNA utilize tracrRNA as an accessory RNA to cleave target DNA sequence that is complimentary to crRNA (12-15).
Since the discovery of tracerRNA in human pathogen Streptococcus pyogenes, computation data reveal the location of tracrRNA on the opposite strand in the upstream from the Cas genes of a Type II-A CRISPR-Cas system (10-15). In fact, tracerRNA is known as a small RNA family that shows absence of sequence similarity within type II CRISPR-Cas loci. Furthermore, tracerRNA has anti-pre-crRNA repeat sequence (anti-repeat) and that enables to play a role in the form trans element (10-15). Due to the presence of inherent anti-repeat sequence in tracerRNA, existence of tracrRNA:pre-crRNA repeat duplexes with pre-crRNA repeats is suggested. Further, these complexes are cleaved by RNase III to activate Cas9 protein (10-15).
Among three types of CRISPR-Cas system, the type II prokaryotic CRISPR-Cas9, an adaptive immune component, shows RNA-guided site-specific DNA cleavage inside genomic loci in various target eukaryotic cells including human cells (1-5). CRISPR-Cas9 system can be integrated with multiple small guided RNAs and this combined are demonstrated to multiple targeting of genes location (6-19).
In a mechanistic landscape, CRISPR-Cas9 system achieves site-specific DNA recognition and cleavage by setting the association of Cas9 nuclease with a CRISPR RNA (crRNA) and additionally trans-activating crRNA (tracrRNA). Essentially, trRNA is discovered to possess complementarity to crRNA and trRNA helps in the maturation of crRNA that can generate multiple copies of pre-crRNAs (6-19).
To use these various types of CRISPR-Cas systems, literature cites potential delivery avenues such as lentiviral vector system, non-viral systems including electroporation, streptavidin-like molecules as a carrier, nanoscale biotechnology and exosome/extracellular vesicle mediated delivery will have future scope in both in vitro and in vivo delivery to achieve successful genome engineering (9-15).
CRISPR-Cas9 genome editing technology is actually an adapted form of powerful and inherent adaptive immune system conferred upon the microorganisms (19-27). This adaptive immune system in microorganisms is in the form of CRISPR-Cas9 that exists in three classes support these microorganisms to eliminate invading viruses. In essence, CRISPR-Cas genome editing technologies are known to encompass various classes of nuclease enzyme such as Cas9, Cas1, Cas3, Cas4 and Cas12a (8-19). Interestingly, a recent discovery reports on a new class of gene editing enzyme as CasX that is claimed as more potent and efficient over Cas9 (15). The encoded nuclease Cas9 creates a complex structure with trcrRNA and crRNA and look for DNA sequences that match the spacer sequence present in CRISPR array loci. Type II systems are suggested as the most significant among three types of CRISPR systems and immensely contribute in genome editing technology (20-22,24-26). In recent, issues of off-target activity of CRISPR systems are reported. To remove these problems, attempts to modify CRISPR system are seen with the help of genomics and bioinformatics tools (8-19). Interestingly, the use of CRISPR editing tool is reported to generate programmable 3D nuclear organization that achieve specific set of gene expression (28).
Currently, CRISPR-Cas9 genome editing methodology is appreciably accepted various human cells lines, animal model system such as mice Mus musculus, Zebrafish Danio Reiro and other model system such as fruit fly Drosophila melanogaster, insect model (10-15). To achieve successful application in human cell lines for the development of specific gene knockout, CRISPR-Cas9 editing technologies involves most important step to design sgDNA with ~20 nt with an approach of using either non-homologous end joining or homology-directed repair system. In such genome editing approach that starts with designing of target and sgDNA and modifications of genes may be achieved within one to two weeks (10-15). Apart from the designing and construction of sgDNA, delivery methods of sgRNAs to target cells can be through an expression cassette encompassing PCR amplicons or sgRNA-expressing plasmids. In case of insect model, the steps involved in the use of CRISPR-Cas9 based genome engineering involves the selection of stage of embryo injection, heteroduplex mobility assay as a screening stage, in vivo evaluation of sgRNA and G0 germline screening of edited insect (10-15).
In recent, CRISPR-Cas9 genome editing tool has received appreciable place in various aspects of science including development of genetic human disease models and gene therapy based treatment of human diseases such as cancer, heart disease, mental illness, single gene disorders and human immunodeficiency virus (HIV) infection (8-19). However, the use of CRISPR-Cas9 editing tool in cancer research and therapeutics is faced with issue of tremendous intra- and inter-tumor heterogeneity for a single type of cancer (20-27). Therefore, it is highly pertinent to know the molecular, genomic and epigenomic landscape of cancer and then precise use of CRISPR-Cas9 genome editing tool to correct the aberrant cancer causing genes such as oncogenes.
Applications and limitations of CRISPR-Cas9 editing technology in cancer therapeutics
Human genome is gifted with 75% of viral and other pathogenic microorganism inserted genes and this conundrum justifies that probability of each individual to get cancer in their life time is up to 1/3 (1-14). Genetic engineering (or simply re-writing existing DNA sequences as choice based system, CRISPR-Cas9 has added a new dimensions to realize the degree and significance of genome editing of genomes from microorganisms to human cells (20-27). It appears to be novel and new tools for genetic and cellular engineers, but these steps are done by primitive microorganisms at their own cost and machinery. In essence, investigators do just by making copy of process adopted by simplest cells. In the process of targeted discovery of genome editing tools, investigators forgot to ask simple question, why nature has given these genome editing tools to simple microorganisms, but not human cells. Actually, it is a balance within the nature and ecosystem that simple organisms are gifted with powerful editing tools and surprisingly complex human cells are blessed with outstanding genomes, but devoid of genome editing tools. On the other hand, it is important to share that microorganisms are lacking strong human adaptive immune system as conferred to human system.
Now by the discovery of CRISPR-Cas9 genome editing tools, investigators intend to keep our complex genome and also the power CRISPR-Cas9 genome editing tools to create designer genome/genes for future survival, existence and therapeutics (20-28). It appears to be in contrast, because nature did not included the power of genome editing tools because our genomes are full of genes probably accumulated by similar type of natural genome editing during evolution of billions of years.
In an elegant paper (29), report that use of CRISPR-Cas9 genome editing tool in a normal and cancer cell can be linked to the status of p53 and ensuing DNA damage response. Hence, it would be good to see the status of p53 gene and their products and its implicated cellular response pathways such as DNA damage response. In short, CRISPR-Cas9 genome editing tool is having huge potentials, but caution and care should be taken to move ahead with its therapeutic use.
Another key finding suggests the precise use of CRISPR-Cas9 editing technology in cancer stem cells over the normal stem cells, as wild type active p53 disrupts the action of editing technology in human pluripotent stem cells (29-35). While, cancer stem cells are known to harbor the mutation and gain of functions of mutant p53 and hence may be a good candidate to use CRISPR-Cas9 editing technology top restore the pro-tumor hallmarks by gene therapy (33). Additionally, emerging evidences point out those cancer stem cells a small pool of tumor microenvironment with p53 mutation and also gain of functions within mutant p53 show less prominent DNA damage response and safeguard mechanisms to genome. Therefore, plausible approach to use CRISPR-Cas9 editing tool in blocking the cancer stem cells by editing oncogenes or bringing onco-suppressors gene to quench its potential so serve as the origin for cancer drug resistance and relapse of cancer.
It is well appreciated idea that the use of CRISPR-Cas9 editing tool needs to be specific and selective based on the genome and epigenomic landscape. Therefore, applications of CRISPR-Cas9 editing tool should be centered upon the efficacy, minimal undesirable outcomes and long term impact in cellular landscape of targeted cells (21). Besides amelioration of oncogenes and tumor suppressors, there is an emerging view to tweak the oncogenic miRNAs by the use of CRISPR/Cas9 editing technology (30). The contribution of CRISPR-Cas9 editing tool is also extended in the cancer immunotherapy to create chimeric antigen receptor-T cells (CART) cells and other anti-cancer immune cells. In a recent attempt, authors claim the potential of CRISPR-Cas9 technology in combination with Cas9-low-molecular-weight protamine (LMWP) nano-carriers to edit the KRAS oncogene in lung cancer (36). To address the important aspects of off target impact of CRISPR-Cas9 editing tool (37) present an elegant study showing the absence of substantial off-target mutations in in vivo model of mouse liver. In essence, this study is a good attempt to remove concerns related to the use of CRISPR-Cas9 editing tool in therapeutic purpose. However, a precise study is the need of hour to encompass the issues of intra- and inter-heterogeneity and use of CRISPR-Cas9 editing tool in the removal of oncogenes and correction of tumor suppressor genes. In case of multi-drug resistance in osteosarcoma cells, disruption of CD44 gene by CRISPR-Cas9 is reported to abrogate the problems of drug resistance (38). A recent paper reports on the rapid screening of therapeutic targets including WRN helicase in several cancer types using CRISPR-Cas9 technology and further this study propose the use of CRISPR-Cas9 to identify genomic lethal targets to achieve cancer therapy success (14). Additionally, CRISPR-Cas9 technology is shown to attenuate elevated level of urokinase plasminogen activator receptor and this inhibition is suggested as option to bring down malignancies.
A recent report indicates on the use of CRISPR-Cas9 technology to bring down the E6 and E7 gene expression in case of high risk HPV mediated cervical cancer patients (39,40). A positive response on chimeric antigen receptor (CAR)-T cells mediated immunotherapeutic response in solid tumors is shown to target PD-1 gene editing by CRISPR-Cas9 (41,42) A recent CRISPR-Cas system that utilizes liposome-encapsulated CRISPR/Cas9 genome editing technology is shown disrupt PD-1 gene in T cells that may have applications in adoptive cancer immunotherapy (43). An in vivo study indicates on the use of lipid-based CRISPR/Cas9 delivery system to target HIF-1α that employs plasmids encoding Cas9 and designed sgRNA targeted to HIF-1α (44). Further, CRISPR/Cas9 system is shown to bring down the HBsAg gene and in turn block the proliferative potential of hepatocellular carcinoma cells (45). Taken together, several reports provide experimental data on the potentials of CRISPR-Cas system as anti-cancer therapeutic approach. However, limitations to use of CRISPR-Cas system are seen in the perspectives of genomic instability, genotypic/epigenomic heterogeneity and undesirable off target (secondary tumors).
Truly, investigators intend to disturb the genomic landscape of nature gifted cellular system and therefore, it may bring inevitable consequences of loss integrity of genome and drifting towards designer genome to destructive genome.
CRISPR-Cas9 and DNA repair landscape of cancer
It is logical to see the appreciable application of CRISPR-Cas9 editing tools in the correction of oncogenes in cancer types with high load of p53 mutation including lung cancer (30-35). Expanding data confirm that the tumor suppressor gene TP53 is mutated in 50% of human cancers including breast, cervical and lung cancer and is referred as the indicator of poor prognosis (32). Other than p53 status amelioration, a clear role of CRISPR-Cas9 editing technology is elaborated in EGFR L858R-positive lung cancer by precisely targeting point mutations as C > G, A > G, and T > G point mutations (46). Here, authors claim the specificity of CRISPR-Cas9 editing technology in cancer over normal cells by using protospacer adjacent motif to differentiate cancer single nucleotide mutation from normal cells. In support of link between CRISPR-Cas9 editing tool and DNA damage response (34), emphasize that use of DNA repair protein inhibitor SCR-7 may be a good option in combinatorial cancer therapeutic to avoid the undesirable consequences of editing technology. Therefore, a reasonable scope is warranted to bring back the wild type features of p53 in a specific type of cancer. Importantly, status of p53 within cancer cells, cancer stem cells and cancer associated immune and stromal cells is also a key features to find the success and failures of CRISPR-Cas9 editing technology.
Conclusions and future perspectives
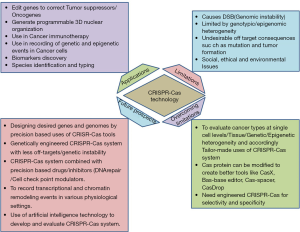
In conclusion, CRISPR associated (Cas) proteins/enzymes, first discovered as Cas9 and then followed by Cas12a, Cas1, Cas4 are highly appreciated as genome editors that use small guided RNAs. In recent, these genome editor tools are faced with issues of off-target impact on the target genome and that leads to undesired mutation and DNA damage response. These unwanted DNA damage responses in the host genome may generate inevitable mutations and potentially a factor for the predisposition to tumor. A caution is also perceived in the use of CRISPR-Cas tools to edit oncogenes and eventually may also trigger the chance of secondary tumors in the host. In the face of these obstacles, Cas proteins/enzymes are continuously modified to create better acceptable tools including CasX, Cas-base editor, Cas-spacer, CasDrop. These Cas variant proteins/enzymes present prospects of CRISPR-Cas genome editor tool, beyond genome editing in case of tumors and other disease condition that include transcriptional and chromatin remodeling. The genome editing potential of CRISPR-Cas9 and other variants of Cas tools is garnering attention in preclinical and clinical success in many types of tumors including oral cancer. Further, the authors opine the use of CRISPR-Cas system in the understanding of basic pathway that contribute to OSCC formation and therapeutic avenues that may bring down the level of key oncogenic driver proteins. Besides these applications in OSCC, oral microbiome in the tumor niche may be targeted by CRISPR-Cas system that acts as pro-tumor agents. Finally, the use modified CRISPR-Cas system may be used in the understanding of transcriptional and chromatin remodeling events that drive the formation of precancerous lesions and initiation of OSCC. The revelation of these molecular, genetic and epigenetic pathways will set the platform for better new classes of therapeutics. A summary of scope, limitation and future modifications in the CRISPR-Cas system is illustrated in Figure 1 (1-15).
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Oral Pre-cancer and Cancer”. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.33). The series “Oral Pre-cancer and Cancer” was commissioned by the editorial office without any funding or sponsorship. SCS served as an unpaid Guest Editor of the series. SP served as an unpaid Guest Editor of the series and serves as an unpaid Editorial Board Member of Translational Cancer Research from Jul 2018 to Jun 2020. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang CH, Lee KC, Doudna JA. Applications of CRISPR-Cas Enzymes in Cancer Therapeutics and Detection. Trends Cancer 2018;4:499-512. [Crossref] [PubMed]
- Fellmann C, Gowen BG, Lin PC, et al. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov 2017;16:89-100. [Crossref] [PubMed]
- Hahn WC. A CRISPR Way to Identify Cancer Targets. N Engl J Med 2019;380:2475-7. [Crossref] [PubMed]
- Kundert K, Lucas JE, Watters KE, et al. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat Commun 2019;10:2127. [Crossref] [PubMed]
- Grünewald J, Zhou R, Garcia SP, et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019;569:433-7. [Crossref] [PubMed]
- Jinek M1. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337:816-21. [Crossref] [PubMed]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819-23. [Crossref] [PubMed]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487-500. [Crossref] [PubMed]
- Pickar-Oliver A, Gersbach CA. The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Biol 2019;20:490-507. [Crossref] [PubMed]
- Nilendu P, Sharma NK. Epigenomic Hard Drive Imprinting: A Hidden Code Beyond the Biological Death of Cancer Patients. J Cancer Prev 2017;22:211-8. [Crossref] [PubMed]
- Gehrke JM, Cervantes O, Clement MK, et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat Biotechnol 2018;36:977-82. [Crossref] [PubMed]
- Mollanoori H, Shahraki H, Rahmati Y, et al. CRISPR/Cas9 and CAR-T cell, collaboration of two revolutionary technologies in cancer immunotherapy, an instruction for successful cancer treatment. Hum Immunol 2018;79:876-82. [Crossref] [PubMed]
- Schmidt F, Cherepkova MY, Platt RJ. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 2018;562:380-5. [Crossref] [PubMed]
- Behan FM, Iorio F, Picco G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019;568:511-6. [Crossref] [PubMed]
- Karimian A, Azizian K, Parsian H, et al. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J Cell Physiol 2019;234:12267-77. [Crossref] [PubMed]
- Liu JJ, Orlova N, Oakes BL, et al. CasX enzymes comprise a distinct family of RNA-guided genome editors. Nature 2019;566:218-23. [Crossref] [PubMed]
- Thomas M, Burgio G, Adams DJ, et al. Collateral damage and CRISPR genome editing. PLoS Genet 2019;15:e1007994. [Crossref] [PubMed]
- Wang K, Xing ZH, Jiang QW, et al. Targeting uPAR by CRISPR/Cas9 System Attenuates Cancer Malignancy and Multidrug Resistance. Front Oncol 2019;9:80. [Crossref] [PubMed]
- Westra ER, van Houte S, Gandon S, et al. The ecology and evolution of microbial CRISPR-Cas adaptive immune systems. Philos Trans R Soc Lond B Biol Sci 2019;374:20190101. [Crossref] [PubMed]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA. recognition by TAL effectors. Science 2009;326:1501. [Crossref] [PubMed]
- Lessard S, Francioli L, Alfoldi J, et al. Human genetic variation alters CRISPR-Cas9 on- and off-targeting specificity at therapeutically implicated loci. Proc Natl Acad Sci U S A 2017;114:E11257-66. [Crossref] [PubMed]
- Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science 2018;361:866-9. [Crossref] [PubMed]
- Sakuma T, Yamamoto T. Acceleration of cancer science with genome editing and related technologies. Cancer Sci 2018;109:3679-85. [Crossref] [PubMed]
- Song CQ, Wang D, Jiang T, et al. In Vivo Genome Editing Partially Restores Alpha1-Antitrypsin in a Murine Model of AAT Deficiency. Hum Gene Ther 2018;29:853-60. [Crossref] [PubMed]
- Urnov FD. Ctrl-Alt-inDel: genome editing to reprogram a cell in the clinic. Curr Opin Genet Dev 2018;52:48-56. [Crossref] [PubMed]
- Wang D, Wang XW, Peng XC, et al. CRISPR/Cas9 genome editing technology significantly accelerated herpes simplex virus research. Cancer Gene Ther 2018;25:93-105. [Crossref] [PubMed]
- Xia AL, He Q, Wang JC, et al. Applications and advances of CRISPR-Cas9 in cancer immunotherapy. J Med Genet 2019;56:4-9. [Crossref] [PubMed]
- Wang H, Xu X, Nguyen CM, et al. CRISPR-Mediated Programmable 3D Genome Positioning and Nuclear Organization. Cell 2018;175:1405-17.e14. [Crossref] [PubMed]
- Haapaniemi E, Botla S, Persson J, et al. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med 2018;24:927-30. [Crossref] [PubMed]
- Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 2018;36:765-71. [Crossref] [PubMed]
- Aquino-Jarquin G. Emerging Role of CRISPR/Cas9 Technology for MicroRNAs Editing in Cancer Research. Cancer Res 2017;77:6812-7. [Crossref] [PubMed]
- Stewart-Ornstein J, Lahav G. p53 dynamics in response to DNA damage vary across cell lines and are shaped by efficiency of DNA repair and activity of the kinase ATM. Sci Signal 2017; [Crossref] [PubMed]
- Chira S, Gulei D, Hajitou A, et al. Restoring the p53 'Guardian' Phenotype in p53-Deficient Tumor Cells with CRISPR/Cas9. Trends Biotechnol 2018;36:653-60. [Crossref] [PubMed]
- Ihry RJ, Worringer KA, Salick MR, et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat Med 2018;24:939-46. [Crossref] [PubMed]
- Solomon H, Dinowitz N, Pateras IS. Mutant p53 gain of function underlies high expression levels of colorectal cancer stem cells markers. Oncogene 2018;37:1669-84. [Crossref] [PubMed]
- Kim SM, Shin SC, Kim EE, et al. Simple in Vivo Gene Editing via Direct Self-Assembly of Cas9 Ribonucleoprotein Complexes for Cancer Treatment. ACS Nano 2018;12:7750-60. [Crossref] [PubMed]
- Akcakaya P, Bobbin ML, Guo JA, et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 2018;561:416-9. [Crossref] [PubMed]
- Xiao Z, Wan J, Nur AA, et al. Targeting CD44 by CRISPR-Cas9 in Multi-Drug Resistant Osteosarcoma Cells. Cell Physiol Biochem 2018;51:1879-93. [Crossref] [PubMed]
- Zhen S, Hua L, Takahashi Y, et al. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun 2014;450:1422-6. [Crossref] [PubMed]
- Yoshiba T, Saga Y, Urabe M, et al. CRISPR/Cas9-mediated cervical cancer treatment targeting human papillomavirus E6. Oncol Lett 2019;17:2197-206. [PubMed]
- Rupp LJ, Schumann K, Roybal KT, et al. CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Sci Rep 2017;7:737. [Crossref] [PubMed]
- Hu W, Zi Z, Jin Y, et al. CRISPR/Cas9-mediated PD-1 disruption enhances human mesothelin-targeted CAR T cell effector functions. Cancer Immunol Immunother 2019;68:365-77. [Crossref] [PubMed]
- Lu S, Yang N, He J. Generation of Cancer-Specific Cytotoxic PD-1- T Cells Using Liposome-Encapsulated CRISPR/Cas System with Dendritic/Tumor Fusion Cells. J Biomed Nanotechnol 2019;15:593-601. [Crossref] [PubMed]
- Li M, Xie H, Liu Y, et al. Knockdown of hypoxia-inducible factor-1 alpha by tumor targeted delivery of CRISPR/Cas9 system suppressed the metastasis of pancreatic cancer. J Control Release 2019;304:204-15. [Crossref] [PubMed]
- Song J, Zhang X, Ge Q. CRISPR/Cas9-mediated knockout of HBsAg inhibits proliferation and tumorigenicity of HBV-positive hepatocellular carcinoma cells. J Cell Biochem 2018;119:8419-31. [Crossref] [PubMed]
- Cheung AH, Chow C, Zhang J, et al. Specific targeting of point mutations in EGFR L858R-positive lung cancer by CRISPR/Cas9. Lab Invest 2018;98:968-76. [Crossref] [PubMed]


