
Role of DNA-dependent protein kinase catalytic subunit in cancer development and treatment
Introduction
The catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) is the key component of the non-homologous end-joining (NHEJ) pathway for DNA double-strand break (DSB) repair and is required for cellular resistance to ionizing radiation (IR) (1,2). DNA-PKcs and the NHEJ pathway are also essential for V(D)J recombination during T and B cell lymphocyte development. Thus, deficiency in DNA-PKcs activity results in a severe combined immunodeficiency (SCID) phenotype in mammals (3-5). In response to DSB formation, DNA-PKcs is recruited to DSBs by the DNA end-binding Ku70/80 heterodimer and is rapidly phosphorylated at multiple serine and threonine residues (1,2). In vivo, DNA-PKcs phosphorylation has been examined at the Thr2609 cluster region and Ser2056. DNA-PKcs phosphorylation at the Thr2609 cluster region is particularly significant since it is critical for DSB repair and radioresistance (6-9). This phosphorylation is regulated by the ataxia telangiectasia mutated (ATM) and the Rad3-related (ATR) kinases in response to various genotoxic stresses, making it a critical regulatory element of DNA-PKcs (6,10). DNA-PKcs phosphorylation at Ser2056, on the other hand, is primarily mediated by autophosphorylation, making it an excellent marker to monitor DNA-PKcs activation in vivo (11).
In addition to its critical function in DSB repair, DNA-PKcs activity has been implicated in cell cycle control; it appears to be involved in mitosis, microtubule dynamics, and proper chromosomal segregation (12). DNA-PKcs activation, as monitored by DNA-PKcs phosphorylation, is physically and functionally associated with mitotic spindle formation. Inhibition of DNA-PKcs activity via a small interfering RNA or a kinase inhibitor results in mitosis delay, abnormal spindle formation, and chromosome misalignment. Furthermore, DNA-PKcs is required to prevent mitotic catastrophe in response to DNA damage (13). These studies unveiled a novel function of DNA-PKcs in safeguarding the genome integrity and cancer suppression as chromosomal instability (CIN) plays an important role in cancer development and is a hallmark of cancer cells (14).
Carcinogenesis is a multi-step process wherein abrogation of multiple cancer susceptibility genes leads to cancer development. Genes that suppress carcinogenesis have been classified as gatekeepers that regulate cellular proliferation and cell death and as caretakers that are primarily encode DNA repair proteins required for the maintenance of genome integrity (15). A direct link between DNA DSBs, genomic instability, and cancer is evidenced the fact that many cancer-predisposition syndromes in humans characterized by genomic instability are caused by mutations in DSB-responsive genes (16,17). In this review, we will discuss our current understanding of the role DNA-PKcs in cancer development and the implications of anti-DNA-PKcs strategies in advanced cancer therapeutics.
The role of DNA-PKcs activity in cancer development
The expression of DNA-PKcs in clinical tumor samples has been investigated to elucidate the impact of DNA-PKcs activity on cancer development. Results generated from such studies not only provide crucial information to predict radio- or chemo-sensitivity of tumor and normal tissues but also help to optimize the treatment plan for each cancer patient. Elevation of DNA-PKcs protein levels has been found in various tumor types by several research groups (Table 1). Significant increases of DNA-PKcs expression levels (protein and mRNA) and kinase activity have been found in colorectal cancers, and these increases are correlated to elevated Sp1 protein levels and poor survival (18,19). High DNA-PKcs protein levels were found in esophageal tumor tissues with an intratumoral heterogeneity pattern compared to the adjacent normal mucosa tissues, where DNA-PKcs expression can be detected in the basal cell layers but not the luminal cells (20). Investigation of nasopharyngeal carcinoma revealed that DNA-PKcs overexpression was found in 36.8% of tumor specimens and had remarkable correlations to advanced clinical stages and poor survival (21). A separate study by Lee et al., however, reported no association between DNA-PKcs overexpression and the clinical outcome of nasopharyngeal carcinoma (22). The difference might be due to the smaller patient volume of the Lee et al. study.
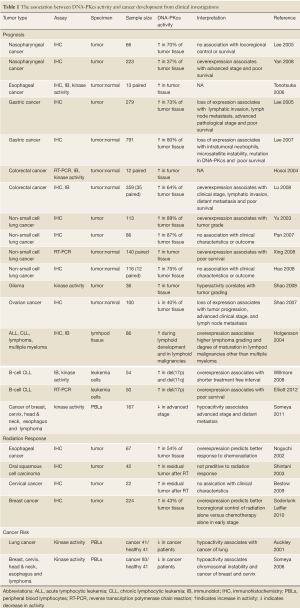
Full table
Elevated mRNA and protein levels of DNA-PKcs as well as several related DSB repair molecules in non small cell lung carcinomas (NSCLCs) have been reported by independent laboratories (23,24). Furthermore, expression profile analysis revealed that patients with higher T/N ratios (tumor samples verse normal tissues) of either DNA-PKcs or ATM have poorer prognosis and increased risk of death; hence, more aggressive therapy is needed (24). This is in agreement with the report by Yu et al. that the high levels of expression of DNA-PKcs protein is associated with tumor grading and expression of p53 and BCL-2, and it might be one of the important causes of radioresistance in NSCLCs (25). There are also reports suggesting no association between DNA-PKcs expression and clinical characteristics or outcome in patients with NSCLC (23,26). Finally, levels of DNA-PKcs activity in glioma specimens coincided with the advanced tumor grading (27). In glioma, DNA-PKcs activity is associated with radioresistance and the epidermal growth factor receptor (EGFR) III variant (VIII) (28), which is among the most mutated genes in glioblastoma multiforme (29,30).
Contrary to the elevated DNA-PKcs activity found in many tumor tissues, the loss of DNA-PKcs expression has been linked to the development of gastric tumors (Table 1). Lee et al. reported that 23% of gastric tumors are negative of DNA-PKcs expression. Lack of DNA-PKcs expression coincides with significant lymphatic invasion, lymph node metastasis, and poor patient survival (31). Further analysis revealed that the loss of DNA-PKcs in advanced gastric tumors is also correlated with intratumoral neutrophils, microsatellite instability, and frame-shift mutations of (A)10 repeats in the DNA-PKcs gene (associated with a higher risk of lymph node metastasis) (32). It is interesting to note that DNA-PKcs is not expressed in epithelium of normal gastric mucosa but is observed in most Helicobacter pylori-associated gastritis, intestinal metaplasia, and gastric adenoma tissues (32). It is possible that the induction of DNA-PKcs is due to hyperproliferation or inflammatory responses triggered by H. pylori. Further studies are needed to clarify the role of DNA-PKcs in H. pylori-induced gastric carcinogenesis (33). Attenuated expression of DNA-PKcs has also been found in ovarian cancer tissue, whereas DNA-PKcs is expressed in 100% of the normal ovaries, in 95% of benign ovary tumors, in 90% in of borderline ovarian neoplasms, and in 60% of malignant serous ovarian neoplasms (34).
Expression of DNA-PKcs and kinase activity in peripheral blood lymphocytes (PBLs) has been examined to correlate individual DNA-PKcs activity to cancer development (Table 1). Studies of PBL samples from cancer patients demonstrated that there is an inverse correlation of DNA-PKcs activity with cancer risk and disease prognosis (35-37). Someya et al. reported that DNA-PK activity is generally attenuated in cancer patients compared to healthy volunteers with statistical significance in breast and cervix cancers (36). Decreased DNA-PKcs activity in PBLs is also found in advanced cancer and is associated with aggressive cancer phenotypes (37). Analyses of advanced cancer patients who have been treated with radiotherapy unveiled a strong correlation between DNA-PKcs activity and the outcome of radiotherapy: Lower DNA-PKcs activity in PBLs was correlated with poorer prognosis including decreased disease-free survival and higher frequency of distant metastasis (37). Additionally, decreased DNA-PKcs activity has also been correlated to chromosomal instability and an increase in chromosomal aberrations (breaks and gaps) in PBLs (36,38). Auckley et al. reported that reduced DNA-PKcs activity in peripheral mononuclear cells (a subset of cells that include PBLs) coincides with lung cancer development (35). Although the authors measured comparable DNA-PKcs activities in PBLs and bronchial epithelial cells (one progenitor cell for lung cancer) of the same individuals, it is unclear whether the coincidence with lung cancer development is due to the reduced DNA-PKcs activity in bronchial epithelial cells or in the immune cells. Nonetheless, these results demonstrate that DNA-PKcs activity in PBLs can be an indicator of aggressiveness of cancer phenotype and patient prognosis.
Comparisons have also been made of expression levels of DNA-PKcs in tumor tissues prior to therapeutic intervention and in the residual tumors after radio- or chemotherapies. High expression of DNA-PKcs was reported to be a predictor for better response to radiation therapy in esophageal cancer and early breast cancer but not in nasopharyngeal cancer (22,39,40). Glioma patients with higher DNA-PKcs activity measured from tumor specimens failed to response to cisplatin-based chemotherapy (27). Conversely, DNA-PKcs and Ku proteins were found to be upregulated after radiation treatment with increased frequency of positive cells in the residual tumors (41,42). The induction of DNA-PKcs and Ku by radiation treatment could be recapitulated in cultured tumor cells suggesting that upregulation of DNA-PKcs and Ku is due to a selective survival advantage against radiation and is associated with radioresistance in the recurrent tumors (41). Thus small molecule inhibitors of DNA-PKcs should be able to sensitize tumors to radiotherapy and facilitate eradication of the radioresistant tumors.
DNA-PKcs activity in lymphoid malignancies
Measurement of DNA-PK activity in PBLs revealed that DNA-PK activity is generally attenuated in cancer patients. The one exception is lymphoma patients who display slightly higher DNA-PKcs activity than the healthy control group (36). Since DNA-PKcs and its downstream NHEJ-dependent DSB repair is essential for V(D)J recombination and lymphocyte development (43,44), it is reasonable to speculate that the increase of DNA-PKcs activity is associated with lymphoid malignancies. Holgersson et al. examined the expression levels of DNA-PK components in different lymphoid malignancies and found that elevated DNA-PKcs and Ku80 protein levels coincide with maturation and high-grade lymphoid malignancies. An increased frequency of DNA-PKcs positive cells was also found in lymph node samples of higher-grade lymphoma patients suggesting that DNA-PKcs is associated with proliferation rate (45). Similar observations were made in patients with B cell chronic lymphocytic leukemia (CLL). Higher levels of DNA-PKcs were found in patients with aggressive types of B cell CLLs (deletions at 17p or 11q) and were correlated to short survival and chemoresistance (46-48). Treatment with DNA-PKcs kinase inhibitor NU7441 sensitizes CLL cells to fludarabine, chlorambucil, and mitoxantrone (47,48). Additionally, increased DNA-PKcs activity was linked to cancer cell survival in acute myeloid leukemia (AML) patients and chemoresistance against various antitumor agents including topoisomerase II (Topo-II) inhibitors (doxorubicin and etoposide) and anti-microtubule vincristine (49). Taken together, these studies demonstrate that DNA-PKcs is an indicator of poor patient prognosis in lymphoid malignancies and may contribute to disease progression. In addition, DNA-PKcs is involved in repair of DNA lesions other than DSBs.
Mutations in the DNA-PKcs gene and cancer development
In addition to the changes in DNA-PKcs activity or expression level, mutations and polymorphisms in DNA-PKcs gene (also referred to as Prkdc or XRCC7) have been examined to explore the possible connection to cancer susceptibility. Several somatic mutations in the coding region of DNA-PKcs have been identified in patients with breast and pancreatic cancers; one missense mutation (c.7825A>C) occurred at the critical Thr2609 phosphorylation cluster of DNA-PKcs (50). DNA-PKcs phosphorylation at the Thr2609 cluster region plays a key role in regulating DNA-PKcs activity and is essential for NHEJ-mediated DSB repair (6-9,51). It is conceivable that the resulting Thr2609Pro mutant DNA-PKcs will interfere with DSB repair, accelerate the accumulation of mutations, and promote genome instability. This notion is further supported by our recent findings that ablation of mouse Thr2605 (human Thr2609) phosphorylation cluster led to multiple tissue stem cells defects and increase of chromosomal aberrations (52). Recently, the first germline mutation of human DNA-PKcs Leu3062Arg was identified from a radiosensitive T-B-SCID patient who also had attenuated Artemis activation and an impaired NHEJ pathway (53). It remains to be determined whether the Leu3062Arg mutation will lead to a tumorigenic phenotype in the patient.
Single nucleotide polymorphism (SNP) analysis revealed that the 6721G to T (rs7003908) variant located in intron 8 of the DNA-PKcs gene is associated with bladder cancer and hepatocellular carcinoma (HCC). Wang et al. reported a dose-dependent and significant increase of bladder cancer risk associated with the 6721T alleles; the risk was profoundly increased in subgroup of 6721TT carriers older than 65 who had smoked (OR=3.24, 95% CI=1.35-7.78) compared with those carrying the 6721 (GG + GT) genotypes (54). Contrary to these findings, Long et al. reported that individuals carrying the homozygote 6721G alleles (GG or GT) have a higher risk of HCC (OR=5.04, 95% CI=3.28-7.76 and OR=3.45, 95% CI=2.40-4.94, respectively) than the 6721TT carriers (55). Additionally, two SNPs in non-coding regions of the DNA-PKcs gene, rs12334811 (G to A, intron 21) and rs8178085 (A to C, intron 29), were associated with decreased risk of lung cancer in a dose-dependent manner (OR=0.53, 95% CI= 0.35-0.80) and significantly in nonsmokers (56). Although the mechanism remains to be determined, these SNPs located in non-coding regions of the DNA-PKcs gene may alter the production levels and/or expression patterns of DNA-PKcs.
Effect of DNA-PKcs inhibitors on cancer cell radiosensitization in vitro
DNA-PKcs and the underlying NHEJ pathway are essential for DSB repair and radioresistance (1,2). As discussed above, elevation of DNA-PKcs activity is associated with tumor radioresistance in some advanced stage tumors. Different anti-DNA-PKcs strategies have been developed to enhance radiotherapy-based control of local tumors or for use in combined modality treatment with other antitumor agents. A number of small-molecule ATP-competitive inhibitors that target DNA-PKcs have been developed including vanillin, SU11752, IC87361, NU7026, and NU7441 Jeny(57-61). Among these, NU7441 is the most potent and selective molecule with a half maximal inhibitory concentration (IC50) of 14 nM against DNA-PKcs in a cell free system and no significant activity against the closely related ATM and ATR kinases (60). All these DNA-PKcs kinase inhibitors have strong radiosensitization effects without causing significant cellular toxicity in various human and murine tissue culture models (57,58,61-65). The presence of a DNA-PKcs inhibitor within the first two hours after radiation exposure, when the majority of DSBs are being repaired, effectively blocks the re-ligation of chromosomal DSBs in a dose-dependent manner; the radiation enhancement ratio of lethal dose to 90% of cells (LD90) ranges from 1.5 to 4.2 in various human cancer cell lines (62). Prolonged incubation with a DNA-PKcs inhibitor further enhanced radiosensitization effects proportionally to exposure time up to 24 hours (63).
DNA-PKcs inhibitors promote IR-induced cell killing primarily through the inhibition of DSB repair and the induction of apoptotic programmed cell death (61,66). Additional mechanisms including acceleration of senescence, induction of mitotic catastrophe, and autophagy have also been implicated in radiosensitization associated with DNA-PKcs kinase inhibition (13,67,68). Azad et al. reported that inhibition of DNA-PKcs induces G2/M checkpoint arrest and an accelerated senescence phenotype in irradiated human NSCLC cells (67). The response of senescence-associated β-galactosidase activity coincides with the induction of p53/p21 signaling pathway and is observed in both tissue culture and tumor xenografts after IR. Recent evidence demonstrates that DNA-PKcs plays a critical role in mitotic spindle formation, proper chromosome segregation, and prevention of mitotic catastrophe in response to DNA damage (12,13). Shang et al. further suggested that DNA-PKcs activity is required for checkpoint kinase 2 (Chk2) activation during the G2/M transition as inactivation of DNA-PKcs results in a prolonged G2/M cell cycle arrest, polyploidy outcome, and higher incidence of multipolar spindles after IR (13). The involvement of DNA-PKcs in autophagy regulation was reported by Zhuang et al. that knockdown of DNA-PKcs radiosensitizes glioma-initiating cells due to IR-induced autophagy responses; expressions of autophagy markers microtubule associated protein light chain 3 (LC3) and Beclin1 are induced as well as the punctate staining patterns of LC3 (autophagosomes) are observed (68). DNA-PKcs-dependent autophagy responses can be blocked by autophagy inhibitor 3-methyladenine, suggesting a role of DNA-PKcs in antagonizing IR-induced autophagy activation. Further analyses are needed to clarify whether the involvement of DNA-PKcs in autophagy responses is a general phenomenon or unique to IR and whether DNA-PKcs kinase activity is required for IR-induced autophagy responses.
Effect of DNA-PKcs inhibition on tumor growth delay in vivo
The effect of DNA-PKcs inhibitors on human cancer cell proliferation has been examined in vivo using xenograft tumor models in mice. There is significant tumor growth delay when mice are treated with combined DNA-PKcs inhibition and IR or Topo-II poisoning (Table 2), and the delay in tumor growth translates into survival benefits (61,62,64,69,70). It is notable that DNA-PKcs inhibition alone does not cause significant antitumor effects and does not cause cytotoxicity. In contrast, the combination of DNA-PKcs inhibition and IR reduces levels of cell proliferation marker Ki67 and increases the signal intensity of the cleaved caspase-3, TUNEL, γH2AX (Ser139), and β-galactosidase in irradiated tumor samples (67,69). In addition to tumor cells, DNA-PKcs inhibition in vivo likely also impacts endothelial cells as greater regression of irradiated tumor vasculature was found in DNA-PKcs-deficient SCID mice and in C57BL6 mice pretreated with DNA-PK inhibitor than in control mice (61). Results generated from these mouse xenograft tumor models further demonstrate the benefit of DNA-PKcs inhibition in sensitizing cells to IR or Topo-II poisons. Thus, anti-DNA-PKcs strategies deserve further investigation in clinical trials.
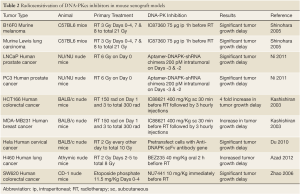
Full table
Additional anti-DNA-PKcs strategies
Additional anti-DNA-PKcs strategies have been developed including single chain antibody variable fragments (scFV) and inhibitory RNA molecules. Anti-DNA-PKcs ScFv 18-2 was derived from an existing anti-DNA-PKcs monoclonal antibody and is able to completely inhibit DNA-PKcs-dependent DNA end joining in vitro (71), whereas Anti-DPK3-scFv was selected from a humanized semi-synthetic scFV library (69). These anti-DNA-PKcs scFVs antagonize DSB repair in vivo and sensitize cells to radiation-induced killing in tissue culture models (69,72,73). Similarly, selective inactivation of DNA-PKcs with inhibitory RNA molecules or antisense oligodeoxynucleotides blocks end-rejoining and sensitizes human cancer cell lines to radiation treatment (70,74,75). Furthermore, studies using mouse xenograft tumor models revealed that both anti-DNA-PKcs scFV (DPK3-scFv) and inhibitory RNAs were able to radiosensitize and to improve local control of tumor growth, thus confirming the antitumor potential of these anti-DNA-PKcs strategies (69,70).
The connection of DNA-PKcs to cancer development has been linked to its association with EGFR, which is frequently overexpressed in human epithelial tumors and is associated with therapy resistance. Wild-type EGFR physically interacts with DNA-PKcs and may modulate the level and activity of DNA-PKcs (76). The interaction of EGFR and DNA-PKcs is also required for IR-induced nuclear AKT phosphorylation and cell survival (77). In NSCLC cell lines expressing EGFR mutated in the tyrosine kinase domain, L858R or △E746-E750, there is impaired EGFR nuclear translocation, no interaction between EGFR and DNA-PKcs, and decreased DNA repair capacity after IR or cisplatin treatment (78,79). Stable exogenous expression of mutant EGFR in human bronchial epithelial cells also abrogates radiation-induced EGFR nuclear translocation and DNA-PKcs binding (78). Blockage of EGFR signaling with a monoclonal antibody or a tyrosine kinase inhibitor could further sensitize cells to radio- or chemotherapies by inhibiting DNA-PKcs activation and decreasing DNA repair capacity (80,81).
Implications of anti-DNA-PKcs strategies in synthetic lethality
Defect in DNA damage repair are often prerequisites to mutation accumulation, genomic instability, and cancer development; as a consequence, cancer cells become addicted to the remains of DNA repair mechanisms to thrive. The synthetic lethality approach exploits this characteristic of cancer cells and selectively targets the remaining DNA repair pathways to kill cancer cells without causing significant cytotoxicity to the normal tissues (82,83). The best example is in breast cancers with mutations in BRCA1 or 2 genes that highly sensitive to inhibitors of poly (ADP-ribose) polymerase 1 (PARP-1) (84). Synthetic lethality is also observed in combined modality therapy of DNA-PKcs inhibitors with other antitumor agents. For example, DNA-PKcs inhibitor NU7026 is able to radiosensitize PARP-1 deficient cells, whereas PARP-1 inhibitor AG14361 radiosensitizes DNA-PKcs-deficient but not PARP-1-deficient cells. NU7026 and AG14361 synergized and reduced levels of recovery of wild-type cells after IR compared to treatment with either agent alone (85,86).
DNA-PKcs inhibitors are also capable of potentiating the cytotoxicity of radiation-mimicking Topo-II poisons including etoposide (VP-16), doxorubicin, and mitoxantrone in human cancer cell lines. This combined modality approach is particularly attractive in treating hematologic (blood) cancers and is effective against most if not all of hematologic cancers including lymphocytic CLL, acute lymphoblastic leukemia, myelocytic chronic myelogenous leukemia and AML, acute promyelocytic leukemia, and adult T-cell leukemia-lymphoma (48,49,87-89). The same strategy is also effective in controlling solid tumors including those of the colon, breast, and prostate (64,65,90). These studies demonstrate that DNA-PKcs and the NHEJ pathway play a crucial role in removing Topo-II poison-induced DSBs that would otherwise lead to caspase activation and apoptosis (91,92).
Increased expression of DNA-PKcs has been linked to cisplatin-resistance in ovarian cancer cells and in glioma patients, suggesting that DNA-PKcs and/or the NHEJ pathway plays a role in DNA interstrand crosslink (ICL) repair (27,49). Consistent with this notion, combined treatment with a DNA-PKcs inhibitor and cisplatin or platinum-based drugs synergize in killing ovarian, colon, and breast cancer cells (57,90,93). In addition to the possible role of DNA-PKcs in ICL lesion repair, DNA-PKcs-mediated cisplatin resistance may occur through AKT activation, EGFR nuclear translocation, or activation/mobilization of chromatin remodeling factor structure-specific recognition protein 1 (SSRP1) from nucleolus (79,94,95). Contrary to the cisplatin resistance model, Jensen and Glazer reported that cells deficient in DNA-PKcs or Ku80 are resistance to cisplatin-induced cell killing through a gap junction-mediated cell death signaling between the neighboring cells (96). This is supported by the observation that decreases in gap junction activity via specific inhibitors or by lowering cell density attenuates cisplatin-induced cell death in DNA-PK-positive cells.
Taken together, these synthetic lethality analyses demonstrate that DNA-PKcs is involved in a variety of DNA damage repair responses in addition to its well-defined role in the NHEJ pathway. We expect that the ongoing and future studies in synthetic lethality screening will unveil other functions of the versatile DNA-PKcs. Additional examples of DNA-PKcs activity and its engagement include resistance to anti-microtubule agent vincristine, DNA intercalator cryptolepine, DNA methylation inhibitor zebularine (49,97,98).
Conclusions and future directions
Current evidence suggests that DNA-PKcs has multiple functions in carcinogenesis and the precise role of DNA-PKcs in cancer promotion or prevention remains to be clarified and may depend on circumstances. The caretaker role of DNA-PKcs in normal tissues promotes DSB repair and chromosomal stability thus preserving and safeguarding the integrity of the genome. Diminished activity of DNA-PKcs thus facilitates accumulation of mutations and genome instability, which has been recognized as a key factor in carcinogenic transformation (99). Elevation of DNA-PKcs expression or kinase activity in the cancer cells reflects the need for greater DNA repair capacity and is likely beneficial for tumor cells given their chronically unstable genomes. Thus, increases in DNA-PKcs activity will enhance cancer cell resistance to DNA damaging and antitumor radio- or chemotherapies and result in the poor patient survival (35,46,100,101). Emerging evidence has also indicated that cancer initiating or cancer stem cells are resistant to radiotherapy and other genotoxic assaults (102,103). Future investigations are needed to determine whether DNA-PKcs plays a role in sustaining cancer stem cell renewal.
The coincidence of diminished DNA-PKcs activity in PBLs and tumor progression suggests that DNA-PKcs activity in PBLs could modulate tumor immunity and initial development of abnormal tumor cells. DNA-PKcs and its downstream NHEJ pathway are essential for V(D)J recombination and the genesis of mature T and B cell lymphocytes (4,5,104). DNA-PKcs has also been implicated in mediating CpG-DNA-dependent macrophage activation and production of IL-10 (105,106). Recent evidence further suggests that DNA-PKcs could play a critical role in the maintenance of hematopoietic stem cells (HSC) and hematogenesis (52) and that DNA-PKcs is required for natural killer (NK) cell activation and the release of pro-inflammatory cytokines (107). Taking into the account that tumor immunity plays a key role in cancer development and progression (108,109), it is possible that the reduction of DNA-PKcs activity in PBLs reflects an overall decrease in host immunity that is essential for suppression of tumor growth or tumor clearance upon radio- or chemotherapeutic interventions. For example, studies using mouse models demonstrate that ablation of NK cells leads to increases in spontaneous tumor development and the frequency of tumor metastasis (108). Future investigations are needed to clarify the function of DNA-PKcs in regulation of macrophage and NK cell activities and tumor immunity.
Acknowledgments
Funding: This work is supported by the National Aeronautics and Space Administration (NNX07AP84G), National Institutes of Health (CA166677), and the Cancer Prevention Research Institute of Texas (RP110465).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2012.04.01). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J 2009;417:639-50. [PubMed]
- Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res 2008;18:114-24. [PubMed]
- Blunt T, Gell D, Fox M, et al. Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc Natl Acad Sci USA 1996;93:10285-90. [PubMed]
- Gao Y, Chaudhuri J, Zhu C, et al. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 1998;9:367-76. [PubMed]
- Kurimasa A, Ouyang H, Dong LJ, et al. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc Natl Acad Sci USA 1999;96:1403-8. [PubMed]
- Chen BP, Uematsu N, Kobayashi J, et al. ATM is essential for DNA-pkcs phosphorylations at T2609 cluster upon DNA double strand break. J Biol Chem 2007;282:6582-7. [PubMed]
- Ding Q, Reddy YV, Wang W, et al. Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol Cell Biol 2003;23:5836-48. [PubMed]
- Nagasawa H, Little JB, Lin YF, et al. Differential role of DNA-PKcs phosphorylations and kinase activity in radiosensitivity and chromosomal instability. Radiat Res 2011;175:83-9. [PubMed]
- Reddy YV, Ding Q, Lees-Miller SP, et al. Non-homologous end joining requires that the DNA-PK complex undergo an autophosphorylation-dependent rearrangement at DNA ends. J Biol Chem 2004;279:39408-13. [PubMed]
- Yajima H, Lee KJ, Chen BP. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol Cell Biol 2006;26:7520-8. [PubMed]
- Chen BP, Chan DW, Kobayashi J, et al. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J Biol Chem 2005;280:14709-15. [PubMed]
- Lee KJ, Lin YF, Chou HY, et al. Involvement of DNA-dependent protein kinase in normal cell cycle progression through mitosis. J Biol Chem 2011;286:12796-802. [PubMed]
- Shang ZF, Huang B, Xu QZ, et al. Inactivation of DNA-dependent protein kinase leads to spindle disruption and mitotic catastrophe with attenuated checkpoint protein 2 Phosphorylation in response to DNA damage. Cancer Res 2010;70:3657-66. [PubMed]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998;396:643-9. [PubMed]
- Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature 1997;386:761, 3.
- O’Driscoll M, Gennery AR, Seidel J, et al. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst) 2004;3:1227-35. [PubMed]
- Jeggo PA. Genomic instability in cancer development. Adv Exp Med Biol 2005;570:175-97. [PubMed]
- Hosoi Y, Watanabe T, Nakagawa K, et al. Up-regulation of DNA-dependent protein kinase activity and Sp1 in colorectal cancer. Int J Oncol 2004;25:461-8. [PubMed]
- Lü Y, Zhang HL, Li YZ, et al. Zhonghua Yi Xue Za Zhi 2008;88:2025-9. [Clinicopathological significance of expressions of DNA dependent protein kinase catalytic subunit and P16 in colorectal carcinoma]. [PubMed]
- Tonotsuka N, Hosoi Y, Miyazaki S, et al. Heterogeneous expression of DNA-dependent protein kinase in esophageal cancer and normal epithelium. Int J Mol Med 2006;18:441-7. [PubMed]
- Yan SS, Liu L, Liu ZG, et al. Expression and clinical significance of DNA-PKcs in nasopharyngeal carcinoma. Ai Zheng 2008;27:979-83. [PubMed]
- Lee S-W, Cho K-J, Park J-H, et al. Expressions of Ku70 and DNA-PKcs as prognostic indicators of local control in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys 2005;62:1451-7. [PubMed]
- Pan H, Zuo C, Mao N, et al. Zhongguo Fei Ai Za Zhi 2007;10:203-5. [Expression and clinical significance of Ku70, Ku80 and DNA-PKcs proteins in patients with stageI-II non-small cell lung cancer by tissue microarray]. [PubMed]
- Xing J, Wu X, Vaporciyan AA, et al. Prognostic significance of ataxia-telangiectasia mutated, DNA-dependent protein kinase catalytic subunit, and Ku heterodimeric regulatory complex 86-kD subunit expression in patients with nonsmall cell lung cancer. Cancer 2008;112:2756-64. [PubMed]
- Yu S, Xiong Y, Tian S. Zhongguo Fei Ai Za Zhi. 2003;6:356-9. [The expression of DNA-PKcs in non-small cell lung cancer and its relationship with apoptosis associated proteins]. [PubMed]
- Hao Y, Hu X, Liu Y, et al. Zhongguo Fei Ai Za Zhi. 2008;11:226-30. [The expression of ERCC1, DNA-PKcs protein and the relation to prognosis in non-small cell lung cancer]. [PubMed]
- Shao CJ, Fu J, Shi HL, et al. Activities of DNA-PK and Ku86, but not Ku70, may predict sensitivity to cisplatin in human gliomas. J Neurooncol 2008;89:27-35. [PubMed]
- Mukherjee B, McEllin B, Camacho CV, et al. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res 2009;69:4252-9. [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [PubMed]
- Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807-12. [PubMed]
- Lee HS, Yang HK, Kim WH, et al. Loss of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) expression in gastric cancers. Cancer Res Treat 2005;37:98-102. [PubMed]
- Lee HS, Choe G, Park KU, et al. Altered expression of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) during gastric carcinogenesis and its clinical implications on gastric cancer. Int J Oncol 2007;31:859-66. [PubMed]
- Gologan A, Graham DY, Sepulveda AR. Molecular markers in Helicobacter pylori-associated gastric carcinogenesis. Clin Lab Med 2005;25:197-222. [PubMed]
- Shao SL, Cai Y, Wang QH, et al. Expression of GLUT-1, p63 and DNA-Pkcs in serous ovarian tumors and their significance. Zhonghua Zhong Liu Za Zhi 2007;29:697-700. [PubMed]
- Auckley DH, Crowell RE, Heaphy ER, et al. Reduced DNA-dependent protein kinase activity is associated with lung cancer. Carcinogenesis 2001;22:723-7. [PubMed]
- Someya M, Sakata K, Matsumoto Y, et al. The association of DNA-dependent protein kinase activity with chromosomal instability and risk of cancer. Carcinogenesis 2006;27:117-22. [PubMed]
- Someya M, Sakata KI, Matsumoto Y, et al. The association of DNA-dependent protein kinase activity of peripheral blood lymphocytes with prognosis of cancer. Br J Cancer 2011;104:1724-9. [PubMed]
- Someya M, Sakata K, Matsumoto Y, et al. Association of DNA-PK activity and radiation-induced NBS1 foci formation in lymphocytes with clinical malignancy in breast cancer patients. Oncol Rep 2007;18:873-8. [PubMed]
- Noguchi T, Shibata T, Fumoto S, et al. DNA-PKcs expression in esophageal cancer as a predictor for chemoradiation therapeutic sensitivity. Ann Surg Oncol 2002;9:1017-22. [PubMed]
- Söderlund Leifler K, Queseth S, Fornander T, et al. Low expression of Ku70/80, but high expression of DNA-PKcs, predict good response to radiotherapy in early breast cancer. Int J Oncol 2010;37:1547-54. [PubMed]
- Shintani S, Mihara M, Li C, et al. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer Sci 2003;94:894-900. [PubMed]
- Beskow C, Skikuniene J, Holgersson A, et al. Radioresistant cervical cancer shows upregulation of the NHEJ proteins DNA-PKcs, Ku70 and Ku86. Br J Cancer 2009;101:816-21. [PubMed]
- Manis JP, Dudley D, Kaylor L, et al. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity 2002;16:607-17. [PubMed]
- Cook AJ, Oganesian L, Harumal P, et al. Reduced switching in SCID B cells is associated with altered somatic mutation of recombined S regions. J Immunol 2003;171:6556-64. [PubMed]
- Holgersson A, Erdal H, Nilsson A, et al. Expression of DNA-PKcs and Ku86, but not Ku70, differs between lymphoid malignancies. Exp Mol Pathol 2004;77:1-6. [PubMed]
- Muller C, Christodoulopoulos G, Salles B, et al. DNA-Dependent protein kinase activity correlates with clinical and in vitro sensitivity of chronic lymphocytic leukemia lymphocytes to nitrogen mustards. Blood 1998;92:2213-9. [PubMed]
- Willmore E, Elliott SL, Mainou-Fowler T, et al. DNA-dependent protein kinase is a therapeutic target and an indicator of poor prognosis in B-cell chronic lymphocytic leukemia. Clin Cancer Res 2008;14:3984-92. [PubMed]
- Elliott SL, Crawford C, Mulligan E, et al. Mitoxantrone in combination with an inhibitor of DNA-dependent protein kinase: a potential therapy for high risk B-cell chronic lymphocytic leukaemia. Br J Haematol 2011;152:61-71. [PubMed]
- Eriksson A, Lewensoh R, Larsson R, et al. DNA-dependent protein kinase in leukaemia cells and correlation with drug sensitivity. Anticancer Res 2002;22:1787-93. [PubMed]
- Wang X, Szabo C, Qian C, et al. Mutational analysis of thirty-two double-strand DNA break repair genes in breast and pancreatic cancers. Cancer Res 2008;68:971-5. [PubMed]
- Chan DW, Chen BP, Prithivirajsingh S, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev 2002;16:2333-8. [PubMed]
- Zhang S, Yajima H, Huynh H, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol 2011;193:295-305. [PubMed]
- van der Burg M, Ijspeert H, Verkaik NS, et al. A DNA-PKcs mutation in a radiosensitive T-B- SCID patient inhibits Artemis activation and nonhomologous end-joining. J Clin Invest 2009;119:91-8. [PubMed]
- Wang SY, Peng L, Li CP, et al. Genetic variants of the XRCC7 gene involved in DNA repair and risk of human bladder cancer. Int J Urol 2008;15:534-9. [PubMed]
- Long XD, Yao JG, Huang YZ, et al. DNA repair gene XRCC7 polymorphisms (rs#7003908 and rs#10109984) and hepatocellular carcinoma related to AFB1 exposure among Guangxi population, China. Hepatol Res 2011;41:1085-93. [PubMed]
- Hu Z, Liu H, Wang H, et al. Tagging single nucleotide polymorphisms in phosphoinositide-3-kinase-related protein kinase genes involved in DNA damage “checkpoints” and lung cancer susceptibility. Clin Cancer Res 2008;14:2887-91. [PubMed]
- Durant S, Karran P. Vanillins--a novel family of DNA-PK inhibitors. Nucleic Acids Res 2003;31:5501-12. [PubMed]
- Ismail IH, Mårtensson S, Moshinsky D, et al. SU11752 inhibits the DNA-dependent protein kinase and DNA double-strand break repair resulting in ionizing radiation sensitization. Oncogene 2004;23:873-82. [PubMed]
- Hollick JJ, Golding BT, Hardcastle IR, et al. 2,6-disubstituted pyran-4-one and thiopyran-4-one inhibitors of DNA-Dependent protein kinase (DNA-PK). Bioorg Med Chem Lett 2003;13:3083-6. [PubMed]
- Leahy JJ, Golding BT, Griffin RJ, et al. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 2004;14:6083-7. [PubMed]
- Shinohara ET, Geng L, Tan J, et al. DNA-dependent protein kinase is a molecular target for the development of noncytotoxic radiation-sensitizing drugs. Cancer Res 2005;65:4987-92. [PubMed]
- Kashishian A, Douangpanya H, Clark D, et al. DNA-dependent protein kinase inhibitors as drug candidates for the treatment of cancer. Mol Cancer Ther 2003;2:1257-64. [PubMed]
- Nutley BP, Smith NF, Hayes A, et al. Preclinical pharmacokinetics and metabolism of a novel prototype DNA-PK inhibitor NU7026. Br J Cancer 2005;93:1011-8. [PubMed]
- Zhao Y, Thomas HD, Batey MA, et al. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res 2006;66:5354-62. [PubMed]
- Shaheen FS, Znojek P, Fisher A, et al. Targeting the DNA double strand break repair machinery in prostate cancer. PLoS ONE 2011;6:e20311 [PubMed]
- Deriano L, Guipaud O, Merle-Béral H, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood 2005;105:4776-83. [PubMed]
- Azad A, Jackson S, Cullinane C, et al. Inhibition of DNA-dependent protein kinase induces accelerated senescence in irradiated human cancer cells. Mol Cancer Res 2011;9:1696-707. [PubMed]
- Zhuang W, Li B, Long L, et al. Knockdown of the DNA-dependent protein kinase catalytic subunit radiosensitizes glioma-initiating cells by inducing autophagy. Brain Res 2011;1371:7-15. [PubMed]
- Du L, Zhou LJ, Pan XJ, et al. Radiosensitization and growth inhibition of cancer cells mediated by an scFv antibody gene against DNA-PKcs in vitro and
in vivo . Radiat Oncol 2010;5:70. [PubMed] - Ni X, Zhang Y, Ribas J, et al. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J Clin Invest 2011;121:2383-90. [PubMed]
- Li S, Takeda Y, Wragg S, et al. Modification of the ionizing radiation response in living cells by an scFv against the DNA-dependent protein kinase. Nucleic Acids Res 2003;31:5848-57. [PubMed]
- Xiong H, Lee RJ, Haura EB, et al. Intranuclear Delivery of a Novel Antibody-Derived Radiosensitizer Targeting the DNA-Dependent Protein Kinase Catalytic Subunit. Int J Radiat Oncol Biol Phys 2012;83:1023-30. [PubMed]
- Xiong H, Li S, Yang Z, et al. coli expression of a soluble, active single-chain antibody variable fragment containing a nuclear localization signal. Protein Expr Purif 2009;66:172-80. [PubMed]
- Sak A, Stuschke M, Wurm R, et al. Selective inactivation of DNA-dependent protein kinase with antisense oligodeoxynucleotides: consequences for the rejoining of radiation-induced DNA double-strand breaks and radiosensitivity of human cancer cell lines. Cancer Res. 2002;62:6621-4. [PubMed]
- Collis SJ, Swartz MJ, Nelson WG, et al. Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res 2003;63:1550-4. [PubMed]
- Bandyopadhyay D, Mandal M, Adam L, et al. Physical interaction between epidermal growth factor receptor and DNA-dependent protein kinase in mammalian cells. J Biol Chem 1998;273:1568-73. [PubMed]
- Toulany M, Kasten-Pisula U, Brammer I, et al. Blockage of epidermal growth factor receptor-phosphatidylinositol 3-kinase-AKT signaling increases radiosensitivity of K-RAS mutated human tumor cells in vitro by affecting DNA repair. Clin Cancer Res 2006;12:4119-26. [PubMed]
- Das AK, Chen BP, Story MD, et al. Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res 2007;67:5267-74. [PubMed]
- Liccardi G, Hartley JA, Hochhauser D. EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res 2011;71:1103-14. [PubMed]
- Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol 2005;76:157-61. [PubMed]
- Friedmann BJ, Caplin M, Savic B, et al. Interaction of the epidermal growth factor receptor and the DNA-dependent protein kinase pathway following gefitinib treatment. Mol Cancer Ther 2006;5:209-18. [PubMed]
- Chan N, Bristow RG. “Contextual” synthetic lethality and/or loss of heterozygosity: tumor hypoxia and modification of DNA repair. Clin Cancer Res 2010;16:4553-60. [PubMed]
- Shaheen M, Allen C, Nickoloff JA, et al. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood 2011;117:6074-82. [PubMed]
- Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol 2011;5:387-93. [PubMed]
- Veuger SJ, Curtin NJ, Richardson CJ, et al. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res 2003;63:6008-15. [PubMed]
- Veuger SJ, Curtin NJ, Smith GCM, et al. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene 2004;23:7322-9. [PubMed]
- Hisatomi T, Sueoka-Aragane N, Sato A, et al. NK314 potentiates antitumor activity with adult T-cell leukemia-lymphoma cells by inhibition of dual targets on topoisomerase II{alpha} and DNA-dependent protein kinase. Blood 2011;117:3575-84. [PubMed]
- Mikusová V, Tichý A, Rezáčová M, et al. Mitoxantrone in Combination with a DNA-PK Inhibitor: Possible Therapy of Promyelocytic Leukaemia Resistant Forms. Folia Biol (Praha) 2011;57:200-5. [PubMed]
- Willmore E, de Caux S, Sunter NJ, et al. A novel DNA-dependent protein kinase inhibitor, NU7026, potentiates the cytotoxicity of topoisomerase II poisons used in the treatment of leukemia. Blood 2004;103:4659-65. [PubMed]
- Davidson D, Grenier J, Martinez-Marignac V, et al. Effects of the novel DNA dependent protein kinase inhibitor, IC486241, on the DNA damage response to doxorubicin and cisplatin in breast cancer cells. Invest New Drugs. 2 2012;30:1736-42.
- Abe T, Ishiai M, Hosono Y, et al. KU70/80, DNA-PKcs, and Artemis are essential for the rapid induction of apoptosis after massive DSB formation. Cell Signal 2008;20:1978-85. [PubMed]
- Friesen C, Uhl M, Pannicke U, et al. DNA-ligase IV and DNA-protein kinase play a critical role in deficient caspases activation in apoptosis-resistant cancer cells by using doxorubicin. Mol Biol Cell 2008;19:3283-9. [PubMed]
- Davidson D, Coulombe Y, Martinez-Marignac VL, et al. Irinotecan and DNA-PKcs inhibitors synergize in killing of colon cancer cells. Invest New Drugs 2012;30:1248-56. [PubMed]
- Dejmek J, Iglehart JD, Lazaro JB. DNA-dependent protein kinase (DNA-PK)-dependent cisplatin-induced loss of nucleolar facilitator of chromatin transcription (FACT) and regulation of cisplatin sensitivity by DNA-PK and FACT. Mol Cancer Res 2009;7:581-91. [PubMed]
- Stronach EA, Chen M, Maginn EN, et al. DNA-PK mediates AKT activation and apoptosis inhibition in clinically acquired platinum resistance. Neoplasia 2011;13:1069-80. [PubMed]
- Jensen R, Glazer PM. Cell-interdependent cisplatin killing by Ku/DNA-dependent protein kinase signaling transduced through gap junctions. Proc Natl Acad Sci USA 2004;101:6134-9. [PubMed]
- Meador JA, Su Y, Ravanat J-L, et al. DNA-dependent protein kinase (DNA-PK)-deficient human glioblastoma cells are preferentially sensitized by Zebularine. Carcinogenesis 2010;31:184-91. [PubMed]
- Zhu H, Gooderham NJ. Mechanisms of induction of cell cycle arrest and cell death by cryptolepine in human lung adenocarcinoma a549 cells. Toxicol Sci 2006;91:132-9. [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [PubMed]
- Townsend DM, Shen H, Staros AL, et al. Efficacy of a glutathione S-transferase pi-activated prodrug in platinum-resistant ovarian cancer cells. Mol Cancer Ther 2002;1:1089-95. [PubMed]
- Harima Y, Sawada S, Miyazaki Y, et al. Expression of Ku80 in cervical cancer correlates with response to radiotherapy and survival. Am J Clin Oncol 2003;26:e80-5. [PubMed]
- Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer 2008;8:545-54. [PubMed]
- Mannino M, Chalmers AJ. Radioresistance of glioma stem cells: intrinsic characteristic or property of the ‘microenvironment-stem cell unit’? Mol Oncol 2011;5:374-86. [PubMed]
- Kirchgessner CU, Patil CK, Evans JW, et al. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science 1995;267:1178-83. [PubMed]
- Dragoi AM, Fu X, Ivanov S, et al. DNA-PKcs, but not TLR9, is required for activation of Akt by CpG-DNA. EMBO J 2005;24:779-89. [PubMed]
- Hazeki K, Kametani Y, Murakami H, et al. Phosphoinositide 3-kinasegamma controls the intracellular localization of CpG to limit DNA-PKcs-dependent IL-10 production in macrophages. PLoS ONE 2011;6:e26836 [PubMed]
- Rajagopalan S, Moyle MW, Joosten I, et al. DNA-PKcs controls an endosomal signaling pathway for a proinflammatory response by natural killer cells. Sci Signal 2010;3:ra14. [PubMed]
- Becknell B, Caligiuri MA. Natural killer cells in innate immunity and cancer. J Immunother 2008;31:685-92. [PubMed]
- Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol 2010;22:231-7. [PubMed]


