
Identification of an individualized autophagy prognostic index in clear cell renal cell carcinoma patients
Introduction
Kidney cancer is a prevalent disease throughout the world, and clear cell renal cell carcinoma (ccRCC) represents the most aggressive and common subtype of this disease (1,2). It has been reported that ccRCC is the most fatal and third most common cancer of the urinary system, which leads to approximately 100,000 deaths every year (3). ccRCC originates from epithelial cells of the renal proximal tubules and is characterized by poor prognosis and high recurrence. As the most common pathological subtype of kidney cancer, the primary treatment option for ccRCC is surgical resection and chemotherapy (4). However, patients who are not suitable for surgery have a low chance of long-term survival. Moreover, few prognostic markers for ccRCC are available in clinical practice due to the influence of a complex network of gene interactions. Thus, it is necessary to identify potential novel prognostic markers and new targets for ccRCC therapy.
Autophagy is an important cellular mechanism which clears damaged organelles and removes abnormal proteins (including aggregated, misfolded and long-lived proteins), to regulate growth and aging (5). Numerous studies have shown that autophagy also plays an essential role in cellular differentiation, nutritional starvation and cellular differentiation (6). Autophagy has an opposing and context-dependent effect on cancer. On one hand, autophagy can prevent the toxic accumulation of mitochondria and damaged proteins, and limit oncogenic signaling, which plays an important role in cancer suppression. On the other hand, tumor cells can use autophagy-mediated recycling to maintain homeostasis, leading to tumor growth and proliferation (7,8). Moreover, autophagy frequently occurs during chemotherapy, and protects tumor cells during treatment resulting in drug resistance and recurrence (9,10). Owing to the complex functions and mechanisms of autophagy, it is necessary to further investigate the association between autophagy and tumors.
The Cancer Genome Atlas (TCGA) was proposed in 2005 as a publicly funded project using genomic analyses to map variations in all human cancers, to better understand the mechanisms of cancer development and progression. We obtained and analyzed datasets including clinical information on ccRCC patients from TCGA database. In particular, we analyzed the expression of autophagy-related genes (ARGs). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of differentially expressed ARGs in ccRCC patients versus healthy controls were performed using Metascape database. Protein-protein networks were constructed using the STRING online database. Hub genes were determined by Cytoscape software. The relationship between the expression levels of hub genes and clinical outcome in ccRCC patients were examined. We developed a prognostic index (PI) as an independent index for overall survival (OS) in ccRCC patients. Finally, the association between PI and clinical factors were investigated.
Methods
Data from the TCGA database
The specific transcriptome expression profiles (FPKM) and corresponding clinical information of ccRCC patients were download from TCGA database. The datasets contained 539 tumor patients and 72 normal controls.
Downloading ARGs
Human autophagy database (HADb) is a web-based public repository containing information about human genes associated with autophagy described thus far. We obtained 234 ARGs from the HADb database.
Differentially expressed ARGs
The R package “limma” was used to identify differentially expressed ARGs between ccRCC patients and normal controls. P value <0.05 and |logFC| >1 were used as cut-off values.
Enrichment analysis of differentially expressed ARGs
GO analysis was used to identify the function of genes, which belonged to three categories: biological processes (BP), cellular components (CC), and molecular functions (MF). KEGG (http://www.genome.jp) is a multi-organism pathway database that contains thousands of pathways and provides specific pathways and linking genomic information. Metascape (http://metascape.org/gp/index.html), a web-based database that provides a comprehensive list of gene annotations and resources for analysis, was used to perform enrichment analyses of differentially expressed ARGs.
Construction and analysis of protein-protein interaction (PPI) network
We used the STRING online database (http://string-db.org) to predict and provide PPI networks after importing differentially expressed ARGs. The open-source Cytoscape software platform was used to provide biological network analysis and two-dimensional (2D) visualization to analyze PPI networks and search hub genes.
Individualized PI based on hub genes
We employed univariate cox regression analyses to identify the hub genes that were significantly associated with OS. multivariate Cox regression analyses were then performed to select genes that could serve as independent indicators in prognostic monitoring. Several central prognostic genes were identified. The PI was calculated as the relative gene expression level multiplied by the relative weight of the genes in the multiple Cox analysis. ccRCC patients were divided into high-risk and low-risk groups using the median PI values as a risk threshold. A log-rank test was performed to assess survival differences between high-risk and low-risk groups, and survival curves were plotted using the Kaplan-Meier (K-M) method.
Statistical analyses
All statistical analyses were conducted using R software (version 3.5.3), GraphPad Prism 7 (San Diego, CA, USA) and SPSS 20.0 (Chicago, IL, USA). The associations between expression profiles of hub genes and OS were analyzed using univariate Cox regression analyses. Based on survival-related factors, a multivariate Cox proportional hazards regression model was used, and PIs were established. The R package “survival ROC” was employed to analyze ROC curves. P values <0.05 were used as cut-off values.
Results
Identification of differentially expressed ARGs
We obtained transcriptional profiles from 539 ccRCC tumor patients and 72 normal controls. A total of 538 ccRCC patients with clinical follow-up information were included. The expression level of 234 ARGs were detected (P value <0.05, |logFC| >1). A total of 43 of these were found to be differentially regulated, comprising 34 up-regulated genes and 9 down-regulated genes (Figure 1A). The heatmap of these genes was shown in Figure 1B and the boxplot of these genes was shown in Figure 1C.
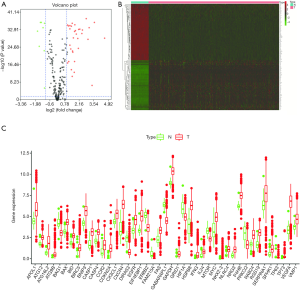
GO and KEGG analysis of the differentially expressed ARGs
GO and KEGG analysis was performed on the 43 identified differentially expressed ARGs. The enrichment results from GO analysis of differentially expressed ARGs found they included the following functions: autophagy, cellular response to external stimulus, autophagosome, apoptotic signaling, ubiquitin protein ligase binding, response to oxygen levels, regulation of cellular response to stress, cellular response to oxidative stress, positive regulation of cell death, selective autophagy, kinase binding, late endosome, vacuolar membrane, positive regulation of organelle organization, anatomical structure homeostasis, cellular response to organonitrogen compound, wound response, regulation of DNA-templated transcription, protein heterodimerization activity, and viral life cycle (Figure 2A,B).
The enrichment results from KEGG analysis were mainly enriched in autophagy (animal), mitophagy (animal), pathways in cancer, apoptosis, longevity regulation, protein processing in the endoplasmic reticulum, hepatitis B, FoxO signaling, tuberculosis, fluid shear stress and atherosclerosis, legionellosis, influenza A, endocytosis, lysosomes, ferroptosis, NF-kappa B signaling, cholinergic synapses, adherens junctions, antigen processing and presentation, and prion diseases (Figure 2C,D).
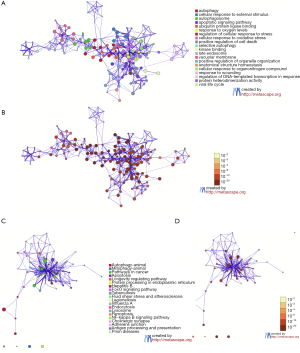
Construction and analysis of the PPI network
The PPI network of DEGs was constructed using the STRING online database and analyzed by Cytoscape software (Figure 3A). The top 25 hub genes were BECN1, SQSTM1, ATG5, ATG12, GABARAPL1, MAP1LC3B, MAP1LC3A, GABARAPL2, ULK1, PINK1, ATG16L1, MTOR, BNIP3, PIK3R4, NBR1, LAMP2, LAMP1, RAB7A, CASP3, BNIP3L, ZFYVE1, GAPDH, MAPK3, MYC, and TP53 (Figure 3B).
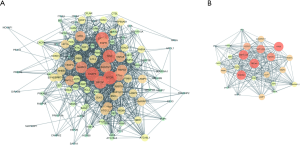
Identification of prognostic hub genes
Univariate Cox regression analyses were employed to explore the relationships between the expression profiles of 25 hub genes and OS in ccRCC patients (Figure 4), which resulted in the identification of prognosis-related ARGs (Figure 4A). To improve credibility, the prognosis-related ARGs were further analyzed using multivariate Cox regression models (Figure 4B). The ARGs with significant prognostic value were BECN1, ULK1, PINK1, BNIP3, and ZFYVE1 (Table 1). Based on the median values of these genes, K-M analysis showed that the up-regulation of ULK1 was closely related to inferior OS rate (Figure 4C,D). On the contrary, up-regulated PINK1, BNIP3, and ZFYVE1 indicated longer survival times.
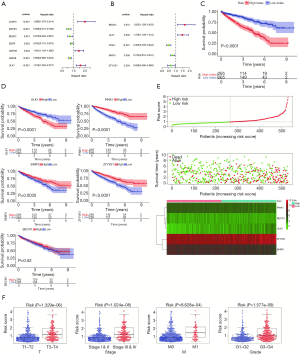
Table 1
| Gene symbol | Description | Gene summary |
|---|---|---|
| BECN1 | Beclin 1 | This gene encodes a protein that regulates autophagy, a catabolic process of degradation induced by starvation. The encoded protein is a component of the phosphatidylinositol-3-kinase (PI3K) complex which mediates vesicle-trafficking processes. This protein is thought to play a role in multiple cellular processes, including tumorigenesis, neurodegeneration and apoptosis. Alternative splicing results in multiple transcript variants (provided by RefSeq, Sep 2015) |
| ULK1 | Unc-51 like autophagy activating kina | GO:2000786 positive regulation of autophagosome assembly; GO:0075044 autophagy of host cells involved in interaction with symbiont; GO:0075071 autophagy involved in symbiotic interaction |
| PINK1 | PTEN induced kinase 1 | This gene encodes a serine/threonine protein kinase that localizes to mitochondria. It is thought to protect cells from stress-induced mitochondrial dysfunction. Mutations in this gene cause one form of autosomal recessive early-onset Parkinson disease (provided by RefSeq, Jul 2008) |
| BNIP3 | BCL2 interacting protein 3 | This gene is encodes a mitochondrial protein that contains a BH3 domain and acts as a pro-apoptotic factor. The encoded protein interacts with anti-apoptotic proteins, including the E1B 19 kDa protein and BCL2. This gene is silenced in tumors by DNA methylation (provided by RefSeq, Dec 2014) |
| ZFYVE1 | Zinc finger FYVE-type containing 1 | The FYVE domain mediates the recruitment of proteins involved in membrane trafficking and cell signaling to phosphatidylinositol 3-phosphate-containing membranes. This protein contains two zinc-binding FYVE domains in tandem and is reported to localize to the Golgi apparatus. Alternative splicing results in multiple transcript variants (provided by RefSeq, Aug 2013) |
ccRCC, clear cell renal cell carcinoma.
Construction and definition of PI
PI was calculated as follows: PI = (0.502576 × expression level of BECN1) + (0.389285 × expression level of ULK1) + (–0.76499 × expression level of PINK1) + (–0.28324 × expression level of BNIP3) + (–0.46899 × expression level of ZFYVE1) (Table 2). According to the median PI, ccRCC patients were divided into high-risk and low-risk groups. The coefficients of PINK1, BNIP3, and ZFYVE1 were negative, while the coefficients of BECN1 and ULK1 were positive. These findings suggest that PINK1, BNIP3, and ZFYVE1 are protective factors, while BECN1 and ULK1 are risk factors.
Table 2
| Gene symbol | Coef | HR | HR.95L | HR.95H | P value |
|---|---|---|---|---|---|
| BECN1 | 0.502576 | 1.652974 | 1.127658 | 2.423007 | 0.010004 |
| ULK1 | 0.389285 | 1.475926 | 1.139319 | 1.91198 | 0.003203 |
| PINK1 | –0.76499 | 0.465339 | 0.336232 | 0.644021 | 3.95e–06 |
| BNIP3 | –0.28324 | 0.753336 | 0.609562 | 0.931022 | 0.008756 |
| ZFYVE1 | –0.46899 | 0.625636 | 0.405697 | 0.964811 | 0.033833 |
| PI = BECN1 × 0.502576 + ULK1 × 0.389285 + PINK1 × (–0.76499) + BNIP3 × (–0.28324) + ZFYVE1 × (–0.46899) | |||||
ARGs, autophagy-related genes; PI, prognostic index.
The ability of PI to predict the clinical outcome of ccRCC patients was analyzed by comparing survival times between the high-risk and low-risk groups by K-M analysis. The results indicated that the survival rate of patients in the high-risk group was significantly worse than that in the low-risk group (Figure 4C). Moreover, when clinicopathological features were adjusted for in multivariate analysis, the PI still represented an independent prognostic indicator (Figure 5A,B). PI distribution in ccRCC patients, number of patients in different risk groups, heat map of five genes, and OS of patients are shown in Figure 4B,C,D,E. Moreover, ROC curve analysis was performed to assess the predictive value of PI for the prognosis of patients with ccRCC. The AUC of PI was 0.811 (Figure 5C).
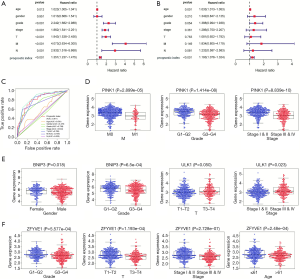
The results of independent sample t-tests indicated that PI was higher in T3–T4 than in T1–T2 (P=1.329e–06), higher in stage III–IV than in I–II (P=1.024e–08), higher in M1 than in M0 (P=8.628e–04), and higher in G3–G4 than in G1–G2 (P=1.577e–09, Figure 4F).
The association between the expression level of these five genes and clinical pathological parameters in ccRCC are shown in Figure 5D,E,F. Low expression of PINK1 had was significantly correlated with advanced pathological M stage (P=2.899e–05), advanced pathological G stage (P=1.414e–08) and advanced pathological stage cancer (P=8.839e–10). Low expression of BNIP3 had a significant correlation with advanced pathological G stage cancer (P=6.5e–04). High expression of ULK1 had a significant correlation with advanced pathological T stage (P=0.05) and advanced pathological stage (P=0.023). Finally, low expression of ZFYVE1 was significantly correlated with advanced pathological G stage (P=5.577e–04), advanced pathological T stage (P=1.193e–04), advanced pathological stage (P=2.728e–07) and increased age (P=2.48e–04).
Discussion
ccRCC is the most common type of renal cancer, with few treatment options and high mortality. In recent years, commendable progress has been made in the detection and treatment of ccRCC. However, reliable clinical parameters that can identify disease progression and survival remain elusive due to the fact that prognosis is influenced by a complex network of gene interactions (11). Autophagy is a major protein degradation process that is highly conserved in eukaryotic cells. It has been reported that autophagy is related to prognosis in patients with many cancer types, such as breast cancer and non-small cell lung cancer, because autophagy may uphold tumor cell metabolism under nutrient limiting conditions to promote cell proliferation (12,13). Furthermore, autophagy may promote invasion and metastasis of cancer cells by many mechanism such as anoikis, tumor dormancy, and epithelial-mesenchymal transition (14,15). Thus, exploration of autophagy may provide new perspectives in ccRCC. Unlike previous studies, our research did not focus on signaling genes; rather, we identified the key prognostic ARGs in ccRCC. Moreover, functional annotation and PPI of ARGs were constructed, all of which may be beneficial for treatment.
In recent years, bioinformatics analysis has been increasingly used to screen and identify potential genetic targets of cancers, owing to progress in high-throughput sequencing and the emergence of comprehensive databases such as TCGA and GEO (16). In our study, we analyzed the transcriptome expression profiles and corresponding clinical information of ccRCC patients in order to identify ARGs, to establish any association with prognosis of patients with ccRCC. A total of 43 differentially expressed ARGs were identified in ccRCC patients compared to healthy controls. KEGG analysis of these genes found they were mainly enriched in “pathways in cancer”. Based on these results, we hypothesized that ARGs play a key role in tumorigenesis. Increasing evidence has shown that tumor formation is a complex and highly regulated multi-step process. In the initial stages of a tumor, autophagy plays a tumor-suppressive role, by inactivation of tumor suppressor genes and activating mutations in oncogenes (17,18). Deletion of autophagy genes has been reported to cause tumorigenesis in certain mouse models (19). However, autophagy becomes a protector of malignant cells after tumor formation, by causing drug resistance and relapse (9,20). Thus, the effect of autophagy on tumor cells is complex and variable across disease progress and tumor type (21).
In our study, we identified five key prognostic ARGs (BECN1, PINK1, ULK1, BNIP3, and ZFYVE1). The protein encoded by BECN1 (beclin 1) is a component of the phosphatidylinositol 3-kinase (PI3K) complex that mediates vesicle transport. It has been reported that BECN1 is a tumor suppressing gene; monoallelic loss of BECN1 promotes cancer development and progression (22). Previous studies have shown that down-regulation of BECN1 predicts poor OS of colon and breast cancer patients (22,23). However, the functional mechanisms of BECN1 in ccRCC remains unknown. PINK1 (PTEN induced kinase 1) encodes a serine/threonine protein kinase that localizes to the mitochondria and is a negative regulator of tumor growth. Mechanistically, PINK1 inhibits reactive oxygen species (ROS) and tumor growth via FOXO3a, a major regulator of oxidative stress and superoxide dismutase 2 (24). BECN1 and PINK1 interact in starvation-induced autophagy (25). ULK1 (unc-51 like autophagy activating kinase 1) encodes a protein that plays a pivotal role in autophagy initiation. Tang et al. showed that NSCLC cell lines show high expression of Ulk1, and Ulk1 is negatively correlated with lung cancer prognosis (26). Further, down-regulation of ULK1 has been found in most breast cancer patients (27). Thus, ULK1 could be a potential target for ccRCC therapy. The mitochondrial protein encoded by BNIP3 (BCL2 interacting protein 3) contains the BH3 domain and acts as a pro-apoptotic factor, which functions as a tumor suppressor during tumorigenesis and as a prognostic indicator of progression to metastasis in certain subtypes of human breast cancer (28). It has been reported that this gene is silenced in tumors by DNA methylation (29). The protein encoded by ZFYVE1 (zinc finger FYVE-type containing 1) contains two FYVE domains bound to zinc, which can mediate proteins involved in membrane trafficking and cell signaling involving phosphatidylinositol 3-phosphate. It has been reported that ZFYVE1 is associated with immunity, which plays a role in some types of cancer (30).
To date, with advancements in large-scale public databases, some prognostic features of cancer have been proposed based on gene expression profiling. For example, Li et al. analyzed RNA-seq data of 119 ccRCC tumors from the GEO database and identified genes significantly associated with OS in ccRCC patients, while Zhang et al. identified 6 that may be associated (31,32). However, these studies focused only on molecular biomarkers, ignoring traditional clinical parameters. In our study, there was a strong emphasis on clinical parameters and molecular mechanisms, to go beyond prognostic features into clinical applications. Moreover, we focused on ARGs to potential targets for ccRCC.
In conclusion, we analyzed transcriptome expression profiles and corresponding clinical information of ccRCC patients, and identified five prognostic ARGs (BECN1, ULK1, PINK1, BNIP3, and ZFYVE1) that may provide potential targets for the treatment of ccRCC. Moreover, a novel risk score PI was constructed according to the expression level of these genes and HR values, which could predict patient survival. Further prospective experiments are warranted to search for optimal personalized targeted therapies and test their clinical utility.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: The authors have completed the ICMJE uniform disclosure from (available at http://dx.doi.org/10.21037/tcr.2020.03.06). The authors have no conflicts of interests to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mehdi A, Riazalhosseini Y. Epigenome aberrations: emerging driving factors of the clear cell renal cell carcinoma. Int J Mol Sci 2017; [Crossref] [PubMed]
- Wettersten HI, Aboud OA, Lara PN Jr, et al. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol 2017;13:410-9. [Crossref] [PubMed]
- Dagher J, Delahunt B, Rioux-Leclercq N, et al. Clear cell renal cell carcinoma: validation of World Health Organization/International Society of Urological Pathology grading. Histopathology 2017;71:918-25. [Crossref] [PubMed]
- Massari F, Di Nunno V, Ciccarese C, et al. Adjuvant therapy in renal cell carcinoma. Cancer Treat Rev 2017;60:152-7. [Crossref] [PubMed]
- Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci 2017;188:53-67. [Crossref] [PubMed]
- Nagar R. Autophagy: a brief overview in perspective of dermatology. Indian J Dermatol Venereol Leprol 2017;83:290-7. [Crossref] [PubMed]
- Mialet-Perez J, Vindis C. Autophagy in health and disease: focus on the cardiovascular system. Essays Biochem 2017;61:721-32. [Crossref] [PubMed]
- Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 2014;20:460-73. [Crossref] [PubMed]
- Chen C, Lu L, Yan S, et al. Autophagy and doxorubicin resistance in cancer. Anticancer Drugs 2018;29:1-9. [Crossref] [PubMed]
- Li YJ, Lei YH, Yao N, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer 2017;36:52. [Crossref] [PubMed]
- López JI, Errarte P, Erramuzpe A, et al. Fibroblast activation protein predicts prognosis in clear cell renal cell carcinoma. Hum Pathol 2016;54:100-5. [Crossref] [PubMed]
- Gu Y, Li P, Peng F, et al. Autophagy-related prognostic signature for breast cancer. Mol Carcinog 2016;55:292-9. [Crossref] [PubMed]
- Liu G, Pei F, Yang F, et al. Role of autophagy and apoptosis in non-small-cell lung cancer. Int J Mol Sci 2017; [Crossref] [PubMed]
- Bhat P, Kriel J, Shubha Priya B, et al. Modulating autophagy in cancer therapy: advancements and challenges for cancer cell death sensitization. Biochem Pharmacol 2018;147:170-82. [Crossref] [PubMed]
- Chen P, Cescon M, Bonaldo P. Autophagy-mediated regulation of macrophages and its applications for cancer. Autophagy 2014;10:192-200. [Crossref] [PubMed]
- Chan LL, Jiang P. Bioinformatics analysis of circulating cell-free DNA sequencing data. Clin Biochem 2015;48:962-75. [Crossref] [PubMed]
- Poillet-Perez L, Despouy G, Delage-Mourroux R, et al. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol 2015;4:184-92. [Crossref] [PubMed]
- Wang RC, Wei Y, An Z, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012;338:956-9. [Crossref] [PubMed]
- Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab 2017;25:1037-43. [Crossref] [PubMed]
- Terabe T, Uchida F, Nagai H, et al. Expression of autophagy-related markers at the surgical margin of oral squamous cell carcinoma correlates with poor prognosis and tumor recurrence. Hum Pathol 2018;73:156-63. [Crossref] [PubMed]
- Bono S, Lulli M, D'Agostino VG, et al. Different BCR/Abl protein suppression patterns as a converging trait of chronic myeloid leukemia cell adaptation to energy restriction. Oncotarget 2016;7:84810-25. [Crossref] [PubMed]
- Peng Y, Miao H, Wu S, et al. ABHD5 interacts with BECN1 to regulate autophagy and tumorigenesis of colon cancer independent of PNPLA2. Autophagy 2016;12:2167-82. [Crossref] [PubMed]
- Tang H, Sebti S, Titone R, et al. Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine 2015;2:255-63. [Crossref] [PubMed]
- Agnihotri S, Golbourn B, Huang X, et al. PINK1 is a negative regulator of growth and the Warburg effect in glioblastoma. Cancer Res 2016;76:4708-19. [Crossref] [PubMed]
- Gelmetti V, De Rosa P, Torosantucci L, et al. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 2017;13:654-69. [Crossref] [PubMed]
- Tang F, Hu P, Yang Z, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol Rep 2017;37:3449-58. [Crossref] [PubMed]
- Zhang L, Fu L, Zhang S, et al. Discovery of a small molecule targeting ULK1-modulated cell death of triple negative breast cancer in vitro and in vivo. Chem Sci 2017;8:2687-701. [Crossref] [PubMed]
- Chourasia AH, Macleod KF. Tumor suppressor functions of BNIP3 and mitophagy. Autophagy 2015;11:1937-8. [Crossref] [PubMed]
- Qu Y, Dang S, Hou P. Gene methylation in gastric cancer. Clin Chim Acta 2013;424:53-65. [Crossref] [PubMed]
- Xu Y, Shen J, Ran Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy 2020;16:3-17. [Crossref] [PubMed]
- Zhang B, Wu Q, Wang Z, et al. The promising novel biomarkers and candidate small molecule drugs in kidney renal clear cell carcinoma: evidence from bioinformatics analysis of high-throughput data. Mol Genet Genomic Med 2019;7:e607. [Crossref] [PubMed]
- Li F, Guo P, Dong K, et al. Identification of key biomarkers and potential molecular mechanisms in renal cell carcinoma by bioinformatics analysis. J Comput Biol 2019;26:1278-95. [Crossref] [PubMed]


