
Microbiome characteristics and Bifidobacterium longum in colorectal cancer patients pre- and post-chemotherapy
Introduction
With the development of next generation sequencing technology, much research has focused on the role of human microbiome in regulating immunity (1,2). Previous studies that involved human and animal models have shown that fluctuations in microbiome levels are associated with the development of various disease states, including metabolic dysfunction, inflammation, infection and cancer (3-7). A number of studies have published their investigations on the risk and clinical outcomes of gastric and colon cancer mediated through the gut microbiome (8-10), and identified various bacterial species that exhibit potential beneficial or pathological effects (11,12). Especially for Actinomyces, a chronic granulomatous disease caused by an anaerobic Gram-positive organism is normally found in the mouth, gut and genitourinary tract, while progressive actinomycosis as well as advancing malignancy may play significant roles in a patient’s demise, although it is a rare but potentially fatal combination of disease and a hitherto undescribed cause of unresectable rectal cancer (13). Breakdown of the mucosal barrier due to trauma (prior to surgery, endoscopic procedures or intestinal perforations, etc.), immunosuppression (steroid therapy, diabetes or tumors) or chronic inflammatory diseases caused by foreign bodies, may be triggered by the penetration of Actinomyces bacteria into the abdomen (14,15). Furthermore, fecal microflora transplantation (FMT) has been shown to reduce the risk of infection in immunocompromised patients (16). However, the optimal composition of microbiota transplants for restoring gut microbiota in colorectal cancer patients undergoing intense anticancer therapies remains uncertain. Furthermore, we are unaware of any study that has closely examined Actinomyces bacteria isolated from the gut microbiota of colorectal cancer patients.
We analyzed the fecal microbiota of colorectal cancer patients who achieved remission following chemotherapy without antimicrobial therapy, and we compared the data obtained before treatment. Bacterial taxa were identified based on 16S ribosomal RNA (rRNA) sequences, which were determined using next-generation sequencing. The diversity and composition of the Actinomyces bacteria in patients were evaluated to characterize differences before and after chemotherapy. We also examined and isolated Actinomyces bacteria in vitro after culturing the microbiota in fecal microbiota samples. Our results provide insight into the optimal composition of gut microbiota transplants for colorectal cancer patients receiving XELOX chemotherapy.
Methods
Patients
We recruited both patients who had not received chemotherapy and those who were given XELOX chemotherapy [capecitabine (Xeloda, Basel, Switzerland, Roche) plus oxaliplatin (ELOXATIN, Paris, France, Sanofi)] in the Department of Oncology, First Affiliation Clinical Center, General Hospital of PLA between November 10, 2017 and December 15, 2018. We also carried out in vitro pure culture experiments on human intestinal Actinomycetes.
Participants meeting any of the following criteria were included: (I) Patients with newly diagnosed intestinal tumor stage IV (colon and rectal cancer); (II) acquired the first time sample was collected from the tumor before surgery or direct treatment; (III) second sampling was assessed after 8 weeks of the first chemotherapy (2 weeks-regimen 3 cycles or 3 weeks-regimen 2 cycles); (IV) XELOX was the main chemotherapy regimen; (V) during second sampling, no severe diarrhea, systemic or intestinal infection, intestinal obstruction, multiple organ failure and other severe diseases were detected.
Exclusion criteria: (I) non-newly diagnosed patients; (II) non-colorectal cancer and (or) non-stage IV intestinal tumors; (III) the first time sample was collected from the patients received surgery or received direct therapy for the treatment; (IV) second sampling time was over 1months of the first evaluation period; (V) main chemotherapy regimens were not XELOX based regimen; and (VI) during the second sample, patients with severe diarrhea, whole body or intestinal infection, intestinal obstruction, accompanied by severe diseases such as multiple organ failure.
According to these selection criteria, only colorectal cancer patients who were III/IV stages who had not underwent antimicrobial therapy before or after treatment with chemotherapy were included in the study. Fecal specimens were collected from patients on the day of enrollment, before the first chemotherapy infusion (baseline) and 8 weeks after completing XELOX chemotherapy. After collection, the stool samples were cryopreserved for subsequent analysis.
Our study was performed in accordance with the principles of the Declaration of Helsinki with regard to ethical research involving human subjects and the protocols were approved by the Medical Ethic Committee of The General Hospital of PLA (approval ID: S2018-081-02). Written informed consent was obtained from all participants prior to enrollment.
Fecal sample collection processing
Each patient was pre-padded with a sterile urine pad and after defecation the central part of the feces was removed from the pad with a sterile spoon and placed in a sterile ice-cold poop box. The sample was subsequently transferred for preservation to a freezer at −80 °C and stored for subsequent analysis. The time of fecal sampling to its storage in a −80 °C freezer was <3 h.
DNA extraction from fecal specimens and 16S rRNA sequencing
DNA was extracted from fecal samples using the QIAamp Power Fecal DNA kit (QIAGEN, German), according to the manufacturer’s instructions, with slight modifications (17). DNA was collected in sterile tubes and the concentration measured by D260/280 with Nanodrop. This method detected genomic DNA purity using 1% agaric gel electrophoresis (100 V, 60 min, with λ-Hind marker III).
A template of the V4−V6 region of the 16S rRNA was generated using universal V4F primer (5'-GTGCCAGCMGCCGCGGTAA-3') universal V6R primer (5'ACAGCCATGCNCACCT-3'). Sequencing library amplicons were generated using barcode primer and reverse primer. Amplicons pools were paired-end sequenced (2×250 bp) in an Illumina MiSeq Sequencer (San Diego, California, USA) at the Beijing Genomics Institute (Beijing, China). After filtering out low quality reads, FLASH version 1.2.11 was used to assemble the paired-end reads, and UCHIME version 4.2.40 was used to remove chimeric sequences before alignment with the Gold Database (ver. 2011.05.19).
Bioinformatics and statistical analysis
Operational taxonomic units (OTUs) were generated from complete linkage clusters using USEARCH ver. 7.0.1090, with a 97% similarity compared to de novo clusters. The OTU were aligned with the Greengenes Database (ver. 2013.05) and annotated for species composition and abundance, as previously described (18). The β diversity at the phylum level was calculated based on the weighted UniFrac distance of rarefied tables (19). We used a linear discrimination analysis (LDA) with effect size (LEfSe) on OTU tables with a LDA score >2.0 to determine the most abundant genera and species in each study group (20). MetaStat analysis was used to evaluate differences in species abundance between the study groups (21). Statistical analysis was conducted using Graphpad Prism ver. 6 (IBM, Armonk, NY, USA). Intergroup differences were evaluated using chi-squared tests or a Kolmogorov-Smirnov test, with the level of significance being set at P<0.05.
Results
Participant characteristics
Four patients were excluded from our analysis. A total of 7 patients [3 female, 4 male, median age: 59 years (range, 42–67 years)] with colorectal cancer were included in our study. The demographic and clinical characteristics of the patients are presented in Table 1. After chemotherapy, the neutrophil (109/L) and platelet counts were significant decreased (0.60±0.12; 174.00±54.87, respectively), compared to pre-treatment. In addition, the expression of several cancer biomarkers was significantly decreased after chemotherapy, except for CA153 (U/L). Serum ferritin was increased in patients in the post-chemotherapy group, which showed that the treatment of patients had been successful with good outcomes (Table 2).
Table 1
| Variable | All patients (n=7) |
|---|---|
| Age [years, median (min, max)] | 59 (42, 67) |
| Gender, n (%) | |
| Male | 4 (57.1) |
| Female | 3 (42.9) |
| Race, n (%) | |
| Han | 7 (100.0) |
| Diet, n (%) | |
| Association with Chinese dietary pattern | 7 (100.0) |
| Tumor location, n (%) | |
| Ascending colon | 1 (14.3) |
| Transverse colon | 2 (28.6) |
| Descending colon | 1 (14.3) |
| Sigmoid colon | 1 (14.3) |
| Rectum | 1 (14.3) |
| Ileocecal cancer | 1 (14.3) |
SD, standard deviation.
Table 2
| Variable | Before treatment (mean ± SD) | After treatment (mean ± SD) | P value |
|---|---|---|---|
| Leucocytes (109/L) | 6.18±2.26 | 5.69±2.86 | 0.728 |
| Neutrophils (109/L) | 13.37±33.79 | 0.60±0.12 | 0.337 |
| Red blood cell distribution width (%) | 12.99±0.70 | 20.41±4.80 | 0.002 |
| Platelets (109/L) | 230.86±41.36 | 174.00±54.87 | 0.049 |
| Thrombin time (sec) | 15.64±1.45 | 15.23±1.13 | 0.566 |
| D-dimer | 4.28±7.14 | 1.43±1.25 | 0.319 |
| Carcinoembryonic antigen (ng/mL) | 92.82±110.05 | 28.86±31.47 | 0.165 |
| CA125 (U/L) | 76.73±82.70 | 41.97±43.74 | 0.345 |
| CA199 (U/L) | 2,992.45±7,504.22 | 661.37±1,687.11 | 0.438 |
| CA153 (U/L) | 12.39±13.79 | 16.09±12.26 | 0.605 |
| CA724 (U/L) | 66.19±89.46 | 24.44±38.87 | 0.280 |
| Cyto-keratin 19 fragment antigen 21 CYFRA21-1 (U/L) | 15.65±17.24 | 3.28±1.19 | 0.083 |
| Neuron-specific enolase (ng/mL) | 15.32±5.30 | 10.39±2.42 | 0.045 |
| Squamous carcinoma-associated antigen (ng/mL) | 1.70±2.62 | 0.79±0.38 | 0.381 |
| Serum ferritin (ìg/L) | 93.10±73.12 | 267.43±324.08 | 0.190 |
| Alanine aminotransferase (U/L) | 15.43±6.67 | 20.33±12.95 | 0.391 |
| Aspartate transaminase (U/L) | 17.39±4.35 | 23.69±5.90 | 0.042 |
| Albumin (g/L) | 38.30±4.22 | 38.93±2.98 | 0.753 |
| Total bilirubin (ìmol/L) | 8.60±2.50 | 11.04±3.79 | 0.181 |
| Direct bilirubin (ìmol/L) | 2.46±0.88 | 3.26±0.91 | 0.120 |
| Total bile acids (ìmol/L) | 2.70±2.35 | 3.95±2.33 | 0.337 |
| Alkaline phosphatase (IU/L) | 139.54±116.09 | 96.19±58.01 | 0.394 |
| γ-Glutamyl transpeptidase (U/L) | 52.63±55.65 | 81.07±128.22 | 0.600 |
| Glucose (mmol/L) | 4.51±0.37 | 5.13±0.88 | 0.111 |
| Serum creatinine (ìmol/L) | 74.13±13.03 | 66.47±12.13 | 0.277 |
| Uric acid (ìmol/L) | 276.91±60.37 | 290.74±103.10 | 0.765 |
| Creatine kinase (U/L) | 59.04±34.95 | 50.61±23.95 | 0.608 |
| Lactic dehydrogenase (U/L) | 163.03±31.37 | 170.44±27.10 | 0.645 |
| Phosphorus (mmol/L) | 1.02±0.10 | 1.06±0.16 | 0.585 |
| Magnesium (mmol/L) | 0.90±0.06 | 0.91±0.04 | 0.720 |
| Calcium (mmol/L) | 3.90±0.46 | 3.95±0.38 | 0.828 |
| Sodium (mmol/L) | 142.90±3.45 | 142.21±1.45 | 0.635 |
| Chloride (mmol/L) | 104.19±2.31 | 104.41±2.63 | 0.871 |
| Carbon dioxide (kPa) | 26.05±1.88 | 27.84±1.40 | 0.066 |
SD, standard deviation.
Metagenomic data and Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolism pathway between pre-and post-chemotherapy for colorectal cancer patients
A total of 57.87 Gb of metagenomic data were obtained from 7 colorectal cancer patients before chemotherapy, which means the average score was 8.27 Gb per individual; a total of 389.24 M high quality “Reads” were obtained, and the average high quality reads obtained for each individual was 55.61 M. whereas a total of 63.23 Gb metagenomic data were obtained from 7 cases of colorectal cancer patients after chemotherapy, with an average of 9.03 Gb per individual; a total of 425.13 M high quality “reads” were obtained, and the average quality of reads per individual was 60.73 M.
From the taxa of the phylum, there were some changes in the relative abundance of intestinal microbes in patients between pre-and post-chemotherapy, and then we further studied the detailed difference in bacteria taxa following phylum, order, family, genus and species taxa. First, the bar plot results showed the pattern of relative abundance of all bacteria isolated from stool samples of patients before or after chemotherapy treatment seemed to be difference (Figure 1), but there was no significant differences in the KEGG pathway between the two groups. Figure 1B shows the top five pathways are two-component system, phenylalanine metabolism, degradation of aromatic compounds, β-lactam resistance and folate biosynthesis.
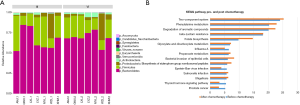
Relative abundances of bacterial kingdom, phylum, class, order, family and species in stool specimens in colorectal cancer patients
In the two groups of samples (pre- or post-chemotherapy), more than 99.6% strains of the intestinal flora were screened out at the level of kingdom, which were bacteria, less than <0.4% were viruses and Archaea (Figure 2A), and then we calculated the relative abundance of the five most common bacterial phylum in the fecal samples (n=7) prior to chemotherapy. As shown in Figure 2B, these top five phylum included Bacteroides, Firmicutes, Proteobacteria, Actinobacteria and Verrucomicrobia. In particular, the abundance of Actinomycetes appeared to be increased after chemotherapy (P=0.078), but for both groups the values did not reach statistical significance.
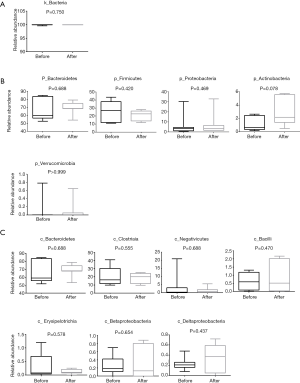
At the level of the class taxa, the relative abundance of Bacteroidia, Clostridia, Negativicute declined in the intestinal flora after chemotherapy. Bacilli, Erysipelotrichia, Betaproteobacteria, Deltaproteobacteria appeared to have increased after chemotherapy (Figure 2C) in the top 20 most abundance of intestinal flora, but there was no significant difference between the pre-and post-chemotherapy groups.
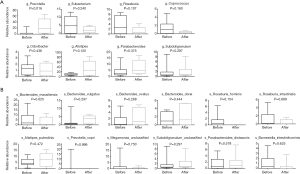
Next, we calculated and compared the top 15 most abundance of intestinal flora in patients before and after chemotherapy at the level of order and family: among them, Bacteroidales, Clostridiales, Selenomonadales, Pasteurellales, Erysipelotrichales, Bifidobacteriales, Lactobacillus, Actinomycetales, Burkholderiales, Desulfovibrionales all exhibited approximate differences in the order level, but only the relative abundance of Bifidobacteriales was significantly increased after chemotherapy.
In addition, at the family level, there seemed to be 8 differences in abundance of intestinal flora in the top 20 family level, namely Prevotellaceae, Lachnospiraceae, Eubacteriaceae, Pasteurellaceae, Erysipelotrichaceae, Ruminococcaceae, Bifidobacteriaceae and Oscillospiraceae (Figure S1A,B), but only Bifidobacteriaceae was significantly increased after chemotherapy.
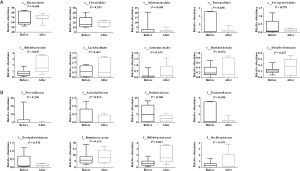
The relative abundances of taxa in the top 20 genus and species before chemotherapy were compared to those of post-chemotherapy at the level of the genus and species, and the results showed significant differences between the study groups (Figure 3). At the level of genes (Figure 3A), the relative abundances of 5 genuses were higher in the pre-chemotherapy colorectal cancer patients and 3 genuses were higher in post-chemotherapy patients, but actually, only Prevotella was statistically significantly increased after chemotherapy. At the level of species (Figure 3B), the relative abundance of 6 species seemed to be higher and 6 species seemed to be lower in the pre-chemotherapy group for the 20 top abundant intestinal flora; these results hinted that colorectal cancer pathology has an impact on Prevotella in the gut microbiome after chemotherapy. The abundance of Prevotella and the severity of postprandial distress-like symptoms in patients with functional dyspepsia exhibited an “inverse correlation” (22).
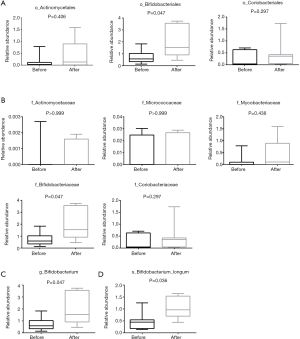
Actinomycetes and Bifidobacterium longum
From the results shown in Figure 2A, at the level of the phylum, the abundance of Actinomycetes appeared to have changed before and after chemotherapy, although the difference was not statistically significant. The abundance of Actinomycetes isolated from patients after chemotherapy treatment was 2.5 fold higher compared to patient levels before chemotherapy, but there was no significant difference of Actinomycetes at the order level. The abundance of Bifidobacteriales at the level of the order and family as well as genus after chemotherapy increased significantly (P=0.047, P=0.047 and P=0.047, respectively) compared to before chemotherapy. A total of 9 genuses were detected for Actinomycetes at the level of the genus during pre- or post-chemotherapy, which included Gordonibacter, Bifidobacterium, Slackia, Collinsella, Actinomyces, Eggerthella, Alloscardovia, Atopobium, At the level of species, 18 were detected to likely have exhibited some changes before and after chemotherapy, but only Bifidobacterium longum was significantly increased and there was a significant difference compared with before chemotherapy (P<0.05) (Figure 4). These findings suggested that there might be a correlation between Bifidobacterium longum species in colorectal cancer, an idea that requires further investigation, while Bifidobacterium longum is a probiotic species that appears to be destined for use in humans (23).
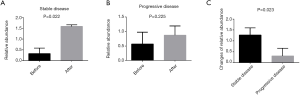
Comparison of the changes of relative abundances of Bifidobacterium longum between patients with favorable (stable disease) and unfavorable outcomes (progressive disease) after chemotherapy from baseline
Based on the above results (supra vide), we further analyzed the relative abundances of Bifidobacterium longum in fecal samples of patients after chemotherapy, and found that there was a significant difference in the relative abundance of Bifidobacterium longum between, after and before chemotherapy in patients with stable disease who achieved more than 6 cycles of the XELOX regimen (P=0.022). There was no significant difference in the relative abundance of Bifidobacterium longum in progressive disease patients (P=0.225). However, we estimate the changes in relative abundance of Bifidobacterium longum species after chemotherapy from baseline in the favorable outcome population (stable disease, SD), was significant higher than in the unfavorable outcome population (progressive disease, PD) (P=0.023) (Figure 5).
Discussion
In the present study, we first confirmed that Bifidobacterium longum, one of the 32 species that belong to the genus Bifidobacterium of the Actinomycetes phylum, was significantly increased in abundance in colorectal cancer patients after XELOX chemotherapy, a finding of great interest.
Actinomycetes are widely found in nature and belong to the bacterial domain (Kingdom) of prokaryotic organisms. The Actinomycetes phylum only contains the Actinomycetes class, which can be divided into six subclasses: Actinobacteria, Acidimicrobiia, Coriobacteriia, Nitriliruptoria, Rubrobacteria, Thermoleophilia, and 8 genus, Actinomycetes Actinomyces, Actinobaculum, Varibaculum, Arcanobacterium, Mobiluncus, Trueperella, Flaviflexus and Falcivibrio (24).
Previous studies have reported that many Actinomycetes strains have rich antibacterial and antitumor activities, and it has been speculated that intestinal Actinomycetes in the human and animals will be a new field for abundant exploitable anti-bacterial resources (25). For example, Bifidobacterium longum, which was increased in abundance in patients after chemotherapy in the present study, has already been confirmed to have many physiological functions beneficial to the human body, and has a strong lipoteichoic acid content in the bacterial cell wall and immunization and antitumor activity in vitro (26). It has also been reported that after chemotherapy in cancer patients, intestinal function is impaired and that the numbers of Bifidobacteria are often significantly reduced (27). However, as normal intestinal flora, Bifidobacterium have the ability to adapt to changes in growth and evolution. Some studies have found that the Bifidobacterium adaptively adjusts both its morphology and resistance to drugs after being inhibited by 5-fluorouracil (5-FU), and successfully overcomes 5-FU inhibition of its growth (28). In a randomized, double-blind, placebo-controlled pilot study of 44 adults with IBS and diarrhea or a diarrhea/constipation-stool pattern and mild to moderate anxiety and/or depression, Bifidobacterium longum NCC3001 showed a benefit in reducing depression (but not anxiety scores) and increasing the quality of life of patients with IBS.
Since the samples of intestinal flora were acquired after chemotherapy in patients with colorectal cancer during the first evaluation period, the interval from the first chemotherapy cycle was nearly 2 months. Thus, it is speculated that the intestinal Bifidobacteria would be inhibited at the early stage of chemotherapy, but may re-grow after adaptive changes and become resistant to chemotherapeutic drugs. In particular, the significant difference in abundance of Bifidobacterium longum at the level of species was observed in patients pre- and post-chemotherapy, which implied that there was an interesting correlation between Bifidobacterium longum and tumors in patients with colorectal cancer, as well as the efficacy of chemotherapy.
Changes in the abundance of Bifidobacteria were found in patients with colorectal cancer to be related to five metabolic functions, which exhibited closely significant differences between the pre-and post-chemotherapy groups, such as glycine, serine and threonine metabolism and xylene degradation, which also need to be further analyzed in future studies.
However, a previous investigation also showed that clinical outcomes for patients undergoing intensive anticancer treatments were influenced by the composition of the gut microbiome (29,30). Therefore, gut microbiota might contribute to the favorable outcome of patients who achieved remission following chemotherapy, without the need for antimicrobial drug treatment. Bifidobacterium longum, a probiotic bacterium with a long list of health benefits for the colon, increased after XELOX chemotherapy. Bifidobacterium longum has anti-inflammatory properties that protect the cells lining mucous membranes from toxins and facilitates immune cells to mature so they can function properly. This probiotic microbe is also present in breast milk and is one of the first microbes to colonize the infant gut (23).
For our pilot study, we recruited patients with colorectal cancer who achieved successful chemotherapy without the need for antimicrobial therapy. We collected stool samples from these patients to examine their fecal microbiota. We characterized differences between the microbiomes of these patients before and after chemotherapy, and we quantified changes in the relative abundances of the colorectal cancer patients’ microbiota produced by chemotherapy. Our results demonstrated that the gut microbiomes of colorectal cancer patients before chemotherapy differed from those post-chemotherapy, although did not exhibit significant different relative abundances in the gut microbiome. Our analysis of colorectal patients before and after chemotherapy showed that the relative abundance of Bifidobacterium longum in stools of colorectal cancer patients was significantly greater than in pre-treatment patients, which suggested that these species might have contributed to favorable outcomes for these patients, since the increase of Bifidobacterium longum in stools of patients with stable disease was significantly higher than in patients with progressive disease (Figure 5).
Previous studies of intensive care unit patients, and patients who received anticancer treatment, revealed that oral and gut microbiota underwent a series of changes upon admission of patients for inpatient care and throughout the course of their treatment, resulting in progressive loss of diversity and enrichment with potentially pathogenic taxa (31-33). A single-center study of intensive care unit patients in Japan found that a ratio of Bacteroidetes to Firmicutes >10 was associated with mortality (34). Bacteroidetes and Firmicutes were among the 10 most abundant phyla in our colorectal cancer patients, but we observed no such extremes in the ratio of these taxa.
The design of the present study did not directly address the effects of the gut microbiome on successful remission of colorectal cancer, but our results do warrant future large-scale clinical investigations of the potential contribution of gut microbiota to remission status in colorectal cancer patients.
Our findings are subject to certain limitations. First, the small sample size of our colorectal cancer patients and second, the single-center nature of our study design limited the statistical power of our findings.
Conclusions
We analyzed the gut microbiomes of colon patients who completed chemotherapy without requiring antimicrobial therapy and had achieved 8 weeks post-treatment survival. Following chemotherapy, the relative abundances of Bifidobacterium longum in stools of colon patients were significantly greater than at pre-chemotherapy. Our results suggested that these taxa might have contributed to the positive clinical outcomes of colon cancer patients. Our results warrant future large-scale multicenter studies to examine microbiota in this small subset of colon patients to determine whether their microbial community structures are associated with favorable clinical outcomes.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.33). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved. Our study was performed in accordance with the principles of the Declaration of Helsinki with regard to ethical research involving human subjects and the protocols were approved by the Medical Ethic Committee of The General Hospital of PLA (approval ID: S2018-081-02). Written informed consent was obtained from all participants prior to enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rogers GB, Bruce KD. Next-generation sequencing in the analysis of human microbiota: essential considerations for clinical application. Mol Diagn Ther 2010;14:343-50. [Crossref] [PubMed]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016;16:341-52. [Crossref] [PubMed]
- Boulangé CL, Neves AL, Chilloux J, et al. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med 2016;8:42. [Crossref] [PubMed]
- Karlsson F, Tremaroli V, Nielsen J, et al. Assessing the Human Gut Microbiota in Metabolic Diseases. Diabetes 2013;62:3341-9. [Crossref] [PubMed]
- Brennan CA, Garrett WS. Gut Microbiota, Inflammation, and Colorectal Cancer. Annu Rev Microbiol 2016;70:395-411. [Crossref] [PubMed]
- Jin C, Lagoudas G, Zhao C, et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019;176:998-1013.e16. [Crossref] [PubMed]
- Kroemer G, Zitvogel L. Cancer immunotherapy in 2017: The breakthrough of the microbiota. Nat Rev Immunol 2018;18:87-8. [Crossref] [PubMed]
- Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 2018;67:226-36. [Crossref] [PubMed]
- Tilg H, Adolph T, Gerner R, et al. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 2018;33:954-64. [Crossref] [PubMed]
- Murphy CL. OʼToole PW, Shanahan F. The Gut Microbiota in Causation, Detection, and Treatment of Cancer. Am J Gastroenterol 2019;114:1036-42. [Crossref] [PubMed]
- Rajagopala SV, Vashee S, Oldfield LM, et al. The Human Microbiome and Cancer. Cancer Prev Res (Phila) 2017;10:226-34. [Crossref] [PubMed]
- Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer 2017;17:271-85. [Crossref] [PubMed]
- Grey T, Lindsay K, Bhowmick A. Actinomycosis: an unusual cause of unresectable rectal cancer. Ann R Coll Surg Engl 2013;95:e92-4. [Crossref] [PubMed]
- Agnihotri M, Kothari K, Naik L. Primary actinomycosis of anterior abdominal wall: A rare occurrence, diagnosed on fine needle aspiration cytology. Indian J Pathol Microbiol 2019;62:629-30. [Crossref] [PubMed]
- Liu K, Joseph D, Lai K, et al. Abdominal actinomycosis presenting as appendicitis: two case reports and review. J Surg Case Rep 2016; [Crossref] [PubMed]
- Kelly CR, Ihunnah C, Fischer M, et al. Fecal Microbiota Transplant for Treatment of Clostridium difficile Infection in Immunocompromised Patients. Am J Gastroenterol 2014;109:1065-71. [Crossref] [PubMed]
- Harata G, Kumar H, He F, et al. Probiotics modulate gut microbiota and health status in Japanese cedar pollinosis patients during the pollen season. Eur J Nutr 2017;56:2245-53. [Crossref] [PubMed]
- Shen X, Miao J, Wan Q, et al. Possible correlation between gut microbiota and immunity among healthy middle-aged and elderly people in southwest China. Gut Pathog 2018;10:4. [Crossref] [PubMed]
- Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl Environ Microbiol 2007;73:1576-85. [Crossref] [PubMed]
- Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [Crossref] [PubMed]
- Wang Z, Wang Q, Wang X, et al. Gut microbial dysbiosis is associated with development and progression of radiation enteritis during pelvic radiotherapy. J Cell Mol Med 2019;23:3747-56. [Crossref] [PubMed]
- Nakae H, Tsuda A, Matsuoka T, et al. Gastric microbiota in the functional dyspepsia patients treated with probiotic yogurt. BMJ Open Gastroenterol 2016;3:e000109. [Crossref] [PubMed]
- Lewis ZT, Shani G, Masarweh CF, et al. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr Res 2016;79:445-52. [Crossref] [PubMed]
- Ji M. Reseaech progress of actinomuces associated clinical infections. J China Pract Diagn Ther 2017;31:815-7.
- Jiang Y, Cao Y, Han L, et al. Diversity and biological activity of pure cultured actinomycetes from five kinds of animal feces. Wei Sheng Wu Xue Bao 2012;52:1282-9. [PubMed]
- Giardina S, Scilironi C, Michelotti A, et al. In vitro anti-inflammatory activity of selected oxalate-degrading probiotic bacteria: potential applications in the prevention and treatment of hyperoxaluria. J Food Sci 2014;79:M384-90. [Crossref] [PubMed]
- Nazir Y, Hussain SA, Abdul Hamid A, et al. Probiotics and Their Potential Preventive and Therapeutic Role for Cancer, High Serum Cholesterol, and Allergic and HIV Diseases. Biomed Res Int 2018;2018:3428437.
- Yuan L, Zhang S, Li H, et al. The influence of gut microbiota dysbiosis to the efficacy of 5-Fluorouracil treatment on colorectal cancer. Biomed Pharmacother 2018;108:184-93. [Crossref] [PubMed]
- Peled JU, Gomes AL, Devlin SM, et al. Inferior survival after microbiota injury: A multicenter allo-HCT study. J Clin Oncol 2019;37:abstr 7015.
- Galloway-Peña JR, Smith DP, Sahasrabhojane P, et al. The role of the gastrointestinal microbiome in infectious complications during induction chemotherapy for acute myeloid leukemia. Cancer 2016;122:2186-96. [Crossref] [PubMed]
- Galloway-Peña JR, Smith DP, Sahasrabhojane P, et al. Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients. Genome Med 2017;9:21. [Crossref] [PubMed]
- Akrami K, Sweeney D. The microbiome of the critically ill patient. Curr Opin Crit Care 2018;24:49-54. [Crossref] [PubMed]
- Rashidi A, Kaiser T, Shields-Cutler R, et al. Outpatient-to-Inpatient Transition Causes Marked Dysbiosis in Allogeneic Hematopoietic Cell Transplantation Recipients. Biol Blood Marrow Tr 2019;25:S47. [Crossref]
- Ojima M, Motooka D, Shimizu K, et al. Metagenomic Analysis Reveals Dynamic Changes of Whole Gut Microbiota in the Acute Phase of Intensive Care Unit Patients. Digest Dis Sci 2016;61:1628-34. [Crossref] [PubMed]


