
MicroRNA-124 and microRNA-378 inhibit the proliferation and invasion of colorectal cancer by upregulating KiSS1
Introduction
Colorectal cancer (CRC) is one of the most common causes of gastrointestinal malignancy and cancer-related death in the clinic (1). Moreover, the number of deaths associated with liver metastases in CRC has exceeded 70% (2). In addition, the overall 5-year survival rate of patients with metastatic CRC in Western countries is only 11%, showing a correlation between the presence or absence of metastasis and the survival of CRC patients (3). Therefore, early detection and treatment are key to improving the prognosis and survival of CRC patients. The incidence and mortality rates of CRC have not yet been significantly reduced because its underlying mechanism is not fully understood. In recent years, with advances in our understanding of tumor molecular biology and the development of genetic engineering, molecular targeted therapy has attracted attention as a new and effective treatment for CRC (4,5).
In an early study of melanoma cell lines, Lee et al. found that the KiSS1 gene is closely related to the occurrence and metastasis of various malignant cancers, including CRC, malignant melanoma, metastatic breast cancer, pancreatic cancer, bladder cancer and gastric cancer (6,7). We have now determined that the KiSS1 gene is located in the chromosome 1q32 region and regulated by genes on chromosome 6 (8). Additionally, the KiSS1 gene is predicted to encode 145 amino acids and can be processed and cleaved into a protein belonging to the Kisspeptin family, including Kisspeptin-10, Kisspeptin-13, Kisspeptin-14, Kisspeptin-54 (9,10). Currently, KISS1 is considered an important tumor suppressor gene in most cancers, but the mechanisms or factors involved in the increased or decreased KiSS1 expression in CRC remain unclear.
MicroRNAs (miRNAs), a class of noncoding single-stranded RNA molecules with a length of approximately 22 nucleotides, are encoded by endogenous genes that are involved in the regulation of posttranscriptional gene expression in plants and animals by inhibiting target mRNA (11). miRNAs are the main regulators of various cell types and are involved throughout the entire cell life cycle, playing important roles in the growth, development, aging, and occurrence of cancer (12). Currently, studies have shown that dysregulated miR-124-3p expression plays certain roles in the occurrence, progression and metastasis of various tumors, including CRC, but the specific mechanism is still unclear (13). Furthermore, miR-378-3p may have an anticancer effect, playing an important role in inhibiting tumor growth and invasion (14). This study was designed to investigate the relationship among miR-124-3p, miR-378-3p and the tumor suppressor gene KiSS1 in CRC.
In our experimental study, we found that miR-124-3p and miR-378-3p upregulate the expression of the tumor suppressor gene KiSS1, thereby inhibiting the proliferation, migration and invasion of CRC cells. In this study, related research on miRNAs and the tumor suppressor gene KiSS1 provides potential and valuable new targets for molecular targeted CRC therapy.
Methods
Cell culture and reagents
The human CRC cell line (SW-480) was cultured in RPMI-1640 medium (HyClone, Beijing, China) supplemented with 10% fetal bovine serum (Gibco Life Technologies, Carlsbad, CA, USA) and 1% penicillin/streptomycin in 95% air and 5% CO2 at 37 °C. SW-480 cells were obtained from the Shanghai Institute of Cell Biology, China Academy of Sciences (Shanghai, China).
Oligonucleotide transfection
To investigate the biological functions of miR-124-3p and miR-378-3p in cell proliferation, migration, invasion and their relationship with KiSS1, miR-124-3p-3p and miR-378-3p-3p mimics were transfected into SW-480 cells to overexpress miR-124-3p and miR-378-3p. In addition, miR-124-3p and miR-378-3p inhibitors were transfected into SW-480 cells to inhibit the expression of miR-124-3p and miR-378-3p. Oligonucleotides including miR-124-3p and miR-378-3p mimics, miR-124-3p and miR-378-3p inhibitor and mimic negative controls and an inhibitor negative control were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). RNA oligonucleotides were transfected at a final concentration of 50 nM using R4000 (Invitrogen, Carlsbad, California, USA) according to the manufacturer’s protocol. To rescue the effects of miR-124-3p and miR-378-3p, a small interfering RNA for KiSS1 (siRNA-KiSS1, 5'-GCCGAACUACAACUGGAACTT-3') and a negative control siRNA (Shanghai GenePharma Co., Ltd. Shanghai, China) were transfected into SW-480 cells. Multiplicities of infection (MOIs) of 10, 20, 40 and 80 were tested in SW-480 cells using lentiviral transfection, and the optimum SW-480 MOI was determined to be 20. After 48 hours of transfection, the harvested cells and supernatant were used in subsequent experiments.
RNA extraction and real-time PCR
Total RNA was extracted from cells cultured from 24 hours under different treatments using TRIzol Reagent (Invitrogen, USA) according to the manufacturer’s protocol. In the presence of an RNase inhibitor (Takara Bio, Shiga, Japan), RNA samples were reverse transcribed using random hexamer primers. Real-time PCR (RT-PCR) was performed on the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA) with SYBR Premix EX Taq (Takara Bio). PCR was performed in a total volume of 20 µL. RT-PCR was performed under the following conditions: 95 °C for 20 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. Relative quantification analysis was performed by the −ΔΔCt method, and β-actin served as the endogenous mRNA reference. Relative gene expression is presented as log(2-ΔΔCt). The following primers were used: KiSS-1 (forward: 5'-AGCCGCCAGATCCCCGCA-3'; reverse: 5'-GCCGAAGGAGTTCCAGTTGTAGTT-3'), β-actin (forward: 5'-CCTCGCCTTTGCCGATCC-3'; reverse: 5'-CCTCGCCTTTGCCGATCC-3'), miR-124-3p (forward: 5'-UAAGGCACGCGGUGAAUGCC-3'; reverse: 5'-GAGCAGGGTCCGAGGT-3'), and miR-378-3p (forward: 5'-GGGACTGGACTTGGAGTCA-3'; reverse: 5'-GTGCGTGTCGTGGAGTCG-3').
Western blot assay
Total protein was extracted from SW-480 cells using a modification buffer containing 0.5% SDS in the presence of a proteinase inhibitor. In total, 60 µg of protein was electrophoresed onto 8% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes. The nonspecific sites were blocked with 5% nonfat milk. The membranes were then incubated with a primary anti-KiSS1 antibody (1:1,000, Abcam) or an anti-GAPDH antibody (1:1,000, Abcam) at 4 °C overnight according to the manufacturer’s instructions and washed three times for 15 min each in Tris-buffered saline with Tween 20 (TBS-T). Next, the blots were incubated for 2 hours at room temperature with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000, Abcam) and washed three times with TBS-T for 15 min. After the final three washes with TBS-T, the blots were visualized on a gel imaging system using Beyo ECL Plus reagent. The relative expression levels of the different proteins were calculated using Bio-Rad Quantity One software.
Cell proliferation assay
Cell proliferation was determined by the 5-ethynyl-20-deoxyuridine (EdU, Beyotime Biotechnology, Shanghai, China) assay in 24-well plates according to the manufacturer's instructions. In total, 1×105 SW-480 cells were seeded in 12-well plates. When the cells were in the logarithmic growth phase, an equal volume of EdU working solution was added to each well. After incubating for 2 hours at 37 °C, the EdU-labeled cells were fixed with 4% paraformaldehyde. After the fixation was complete, 0.5 mL of the Click reaction solution was added to each well. To measure the proportion of proliferating cells, 1 ml of Hoechst 33342 solution was added to each well, and the mixture was incubated for 10 min at room temperature in the dark. Fluorescence was then detected; Hoechst 33342 stains the nuclei of all cells blue, and EdU stains the nuclei of proliferating cells red. The images were taken and analyzed using a digital microscope system (CX41, Olympus).
Cell migration and invasion assays
For the cell migration assay, SW-480 cells under different treatments were seeded in the upper chambers of Transwell units with an 8-mm-pore size polycarbonate filter in 0.5% FBS medium. The lower chamber was filled with 700 µL of RPMI-1640 medium containing 1% FBS. After 24 hours of incubation, the culture was removed, and the filter was fixed with 4% paraformaldehyde for 20 min. Then, the cells on the upper surface of the filter were completely removed with a cotton swab, and the filter was dyed with 0.1% crystal violet for 15 min. Cells that migrated through the upper chamber to the lower surface of the filter were counted and analyzed with a digital microscope system. The experiments were repeated three times.
For the cell invasion assay, the invasive ability of SW-480 cells was determined by seeding cells into the upper chambers of BSA-coated Transwell units with an 8 µM pore size (Corning Star, Cambridge, MA, USA). The cells were then incubated at 37 °C for 24 hours to allow migration through the porous membrane. The remaining cells were completely removed from the upper surface of the chamber, and the filter was then dyed with 0.1% crystal violet. The results were observed using an Olympus CX41 microscope, and the cell numbers in the different treatment groups were determined using Image-Pro Plus 6.0 software.
Statistical analyses
The data were analyzed by ANOVA with Dunnet post hoc test and P<0.05 was considered statistically significant.
Results
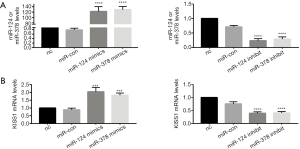
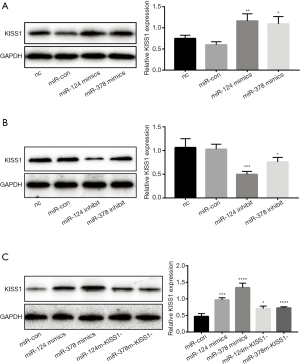
Overexpression of miR-124-3p and miR-378-3p increased KiSS1 mRNA expression. After transfecting with mimics and inhibitors, successful overexpression or inhibition of miR-124-3p and miR-378-3p was determined by RT-qPCR analysis (Table 1, Figure 1A). And transfecting miR-124-3p mimics and miR-378-3p mimics increased KiSS1 mRNA expression (Figure 1B). The results of Western blotting clearly showed that the transfection with miR-124-3p and miR-378-3p mimics significantly increased the synthesis of KiSS1 (Figure 2A). Moreover, the expression of KISS1 mRNA and protein was exactly opposite to that of the mimics group in the cells transfected with miR-124-3p and miR-378-3p inhibitors (Figures 1B,2B). In addition, KISS1 was knocked down on the basis of overexpression of miR-124-3p and miR-378-3p, and the expression of KISS1 was found to be inhibited (Figure 2C).
Table 1
| Primer | Sequence(5'-3') |
|---|---|
| KiSS-1 | |
| Forward | 5'-AGCCGCCAGATCCCCGCA-3' |
| Reverse | 5'-GCCGAAGGAGTTCCAGTTGTAGTT-3' |
| β-actin | |
| Forward | 5'-CCTCGCCTTTGCCGATCC-3' |
| Reverse | 5'-CCTCGCCTTTGCCGATCC-3' |
| miR-124-3p | |
| Forward | 5'-UAAGGCACGCGGUGAAUGCC-3' |
| Reverse | 5'-GAGCAGGGTCCGAGGT-3' |
| miR-378-3p | |
| Forward | 5'-GGGACTGGACTTGGAGTCA-3' |
| Reverse | 5'-GTGCGTGTCGTGGAGTCG-3' |
Overexpression of miR-124-3p and miR-378-3p repressed the proliferation of SW-480 cells in vitro. As shown in Figure 3A, after transfection with the mimics, the percentage of EdU-positive cells, which indicated the number of proliferating cells, decreased; in addition, the proliferation ability of SW-480 cells increased after transfection with miR-124-3p and miR-378-3p inhibitors (Figure 3A).
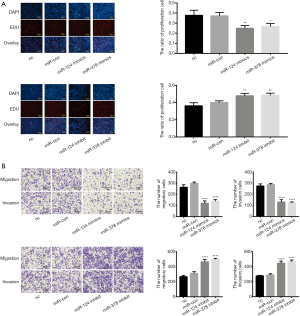
Overexpression of miR-124-3p and miR-378-3p decreased the invasion and migration abilities of SW-480 cells. Overexpression of miR-124-3p and miR-378-3p significantly inhibited the migration and invasion of SW-480 cells, and inhibition of miR-124-3p and miR-378-3p significantly attenuated this effect (Figure 3B).
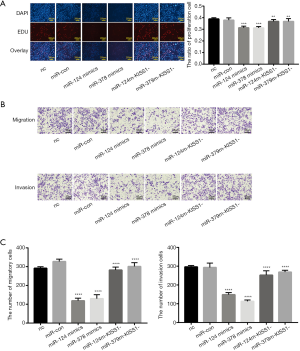
To determine the mechanism by which miR-124-3p and miR-378-3p suppress migration and invasion, we first examined whether miR-124-3p and miR-378-3p could upregulate the expression of the antimetastatic gene KiSS1. We investigated the effect of miR-124-3p and miR-378-3p on KiSS1 by detecting the synthesis of KiSS1. Via cotransfection with siRNA-KiSS1, the proliferation, migration and invasion suppressed cell by miR-124-3p and miR-378-3p overexpression were attenuated (Figure 4). Our results described above indicate that the reduced invasion of CRC cells may be due to miR-124-3p and miR-378-3p and the KiSS1 signal transduction pathway.
Discussion
The pathogenesis of CRC is not fully understood, and infinite proliferation and distant metastasis are the main causes of treatment failure or recurrence. miRNAs have been hotspots in molecular biology research in recent years, and investigators hope that miRNAs can regulate the expression of genes that play roles in cancer and facilitate biological changes to halt tumor growth (15).
In recent years, miR-124-3p has been shown to be expressed at lower levels in medulloblastoma tissues than in normal tissues (16); in addition, studies have reported that methylated miR-124-3p can be used as a potential indicator for cervical cancer screening (17). Furthermore, the prognosis of CRC patients was shown to be related to the level of miR-124-3p expression (18). Moreover, miR-378-3p was shown to inhibit hepatocyte proliferation in mouse liver regeneration experiments (19); miR-378-3p may be a factor in the molecular mechanism by which metformin inhibits liver cancer (20). The efficacies of miR-124-3p and miR-378-3p in tumors have gradually become a topic of focus.
KiSS1 can inhibit tumor growth and metastasis. In CRC, methylation of the KiSS1 gene promoter affects the expression of KiSS1 and thus affects tumor invasion and migration (21). In addition, KiSS1 can also regulate the proliferation, invasion and migration of CRC cells via the PI3K/AKT/NF-κB signaling pathway (22). Moreover, miR-124-3p is also associated with the AKT/GSK-3β/SNAIL-1 signaling pathway in osteosarcoma (23). Direct relationships beyond those currently known may exist between miRNAs and the KiSS1 gene, and these relationships may influence tumor formation through the same or similar cell signaling pathways. In this study, the expression levels of miR-124-3p and miR-378-3p were shown to be upregulated in CRC cells through a series of cytological experiments; in addition, expression of the tumor suppressor gene KiSS1 was detected and found to be significantly increased in CRC cells. On this basis, we continued to detect related cell cytokines and found that the proliferation, migration and invasion of CRC cells transfected with miR-124-3p and miR-378-3p were lower than those in the untransfected group. Our experimental group has been working on the role of KISS1 gene in the development of colorectal cancer in the early stage and found it to be a tumor suppressor gene. In order to further study the related molecular mechanisms affecting KISS1 gene expression, we screened a part of the tumor suppressor microRNA through the miRbase database, and then found that transfection of miR-124-3p and miR-378-3p up-regulated the expression of KISS1 gene. However, whether KiSS1 is a target gene of miR-124-3p and miR-378-3p, and the specific mechanism by which miR-124-3p and miR-378-3p regulate KISS1 is unclear, and perhaps there is an intermediate signaling molecule involved between them. Currently, the mechanisms by which miR-124-3p and miR-378-3p affect the KiSS1 gene and changes in CRC cell biological behaviors remain unknown. Numerous types of miRNAs exist, and these noncoding RNAs do not regulate individual genes. Due to limitations in experimental funding and experimental conditions, the limitations of this study were that no clinical samples were collected and tested in vivo. We will continue to explore the possible roles of miRNAs and the tumor suppressor gene KiSS1 in the development of tumors, including CRC.
Based on the above conclusions, miR-124-3p and miR-378-3p increase the expression of the KiSS1 gene and inhibit the proliferation, migration and invasion of CRC cells. We hope that the results presented herein will provide new ideas for further research on the molecular mechanisms underlying the occurrence of CRC.
Acknowledgments
Funding: The present study was conducted at The First Affiliated Hospital of Fujian Medical University and was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.02.30). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res 2018;30:1-12. [Crossref] [PubMed]
- Neo J, Ager E, Angus P, et al. Changes in the renin angiotensin system during the development of colorectal cancer liver metastases. BMC Cancer 2010;10:134. [Crossref] [PubMed]
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Yeung Y, Tebbutt N. Bevacizumab in colorectal cancer: current and future directions. Expert Rev Anticancer Ther 2012;12:1263-73. [Crossref] [PubMed]
- Khan K, Cunningham D, Chau I. Targeting Angiogenic Pathways in Colorectal Cancer: Complexities, Challenges and Future Directions. Curr Drug Targets 2017;18:56-71. [Crossref] [PubMed]
- Lee J, Miele M, Hicks D, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J. Natl. Cancer Inst 1996;88:1731-7. [Crossref] [PubMed]
- Steeg P. Metastasis suppressors alter the signal transduction of cancer cells. Nat. Rev. Cancer 2003;3:55-63. [Crossref] [PubMed]
- Sanchez-Carbayo M, Capodieci P, Cordon-Cardo C. Tumor suppressor role of KiSS-1 in bladder cancer: loss of KiSS-1 expression is associated with bladder cancer progression and clinical outcome. Am. J. Pathol. 2003;162:609-17. [Crossref] [PubMed]
- Bilban M, Ghaffari-Tabrizi N, Hintermann E, et al. Kisspeptin-10, a KiSS-1/metastin-derived decapeptide, is a physiological invasion inhibitor of primary human trophoblasts. J Cell Sci 2004;117:1319-28. [Crossref] [PubMed]
- Stafford L, Xia C, Ma W, et al. Identification and characterization of mouse metastasis-suppressor KiSS1 and its G-protein-coupled receptor. Cancer Res 2002;62:5399-404. [PubMed]
- Chen C, Li L, Lodish H, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004;303:83-6. [Crossref] [PubMed]
- Ryan BM. microRNAs in Cancer Susceptibility. Adv Cancer Res 2017;135:151-71. [Crossref] [PubMed]
- Yu J, Sun N, Bei Y, et al. Circadian gene hCLOCK contributes to progression of colorectal carcinoma and is directly regulated by tumor suppressive microRNA124. Mol Med Rep 2017;16:7923-30. [Crossref]
- Zhang GJ, Zhou H, Xiao HX, et al. MiR-378 is an independent prognostic factor and inhibits cell growth and invasion in colorectal cancer. BMC Cancer 2014;14:109. [Crossref] [PubMed]
- Lian B, Yang D, Liu Y, et al. miR-128 Targets the SIRT1/ROS/DR5 Pathway to Sensitize Colorectal Cancer to TRAIL-Induced Apoptosis. Cell Physiol Biochem 2018;49:2151-62. [Crossref] [PubMed]
- Pierson J, Hostager B, Fan R, et al. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol 2008;90:1-7. [Crossref] [PubMed]
- Wilting SM, van Boerdonk RA, Henken FE, et al. Methylation-mediated silencing and tumour suppressive function of hsa-miR-124 in cervical cancer. Mol Cancer 2010;9:167. [Crossref] [PubMed]
- Wang MJ, Li Y, Wang R, et al. Downregulation of microRNA-124 is an independent prognostic factor in patients with colorectal cancer. Int J Colorectal Dis 2013;28:183-9. [Crossref] [PubMed]
- Song G, Sharma AD, Roll GR, et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 2010;51:1735-43. [Crossref] [PubMed]
- Zhou J, Han S, Qian W, et al. Metformin induces miR-378 to downregulate the CDK1, leading to suppression of cell proliferation in hepatocellular carcinoma. Onco Targets Ther 2018;11:4451-9. [Crossref] [PubMed]
- Chen S, Chen Z, Lin S, et al. KISS1 methylation and expression as predictors of disease progression in colorectal cancer patients. World J Gastroenterol 2014;20:10071-81. [Crossref] [PubMed]
- Chen S, Chen W, Zhang X, et al. Overexpression of KiSS-1 reduces colorectal cancer cell invasion by downregulating MMP-9 via blocking PI3K/Akt/NF-κB signal pathway. Int J Oncol 2016;48:1391-8. [Crossref] [PubMed]
- Yu B, Jiang K, Zhang J. MicroRNA-124 suppresses growth and aggressiveness of osteosarcoma and inhibits TGF-β-mediated AKT/GSK-3β/SNAIL-1 signaling. Mol Med Rep 2018;17:6736-44. [PubMed]


