Nomograms to predict overall and cancer-specific survival in patients with penile cancer
Introduction
Penile cancer (PC) is a relatively rare malignancy with 2080 estimation new cases in 2019 in the United States, representing 0.24% of all men's new cancer cases (1). The incidence and mortality of PC in developed countries have been low and stable (1-3). However, in South America, Asia, and parts of Africa, the morbidity is much higher, accounting for up to 1–2% of malignant tumors in men (4). In some poor areas, the incidence is as high as (6−7)/100,000 (5). The present paradigm for PC management includes pathological biopsy and immediate excision for highly suspicious lesions (6). Early resection is indeed beneficial to survival, unfortunately, patients who undergo inguinal lymphadenectomy are more likely to short- and long-term morbidity (7,8). In addition, the effects of chemotherapy and radiotherapy remained controversial because of their depressing results (9,10).
As far as we known, prognostic factors such as higher histological grade (11), growth pattern (superficial) (12), perineural invasion (13), venous and/or lymphatic embolization (14), marital status (15), age, race, tumor size, and treatment can influence patient outcomes. For instance, inguinal lymph node involvement is the most important prognostic factor and surgical management of the inguinal region is needed even when the clinical disease is absent (16). However, Lopes found that the T stage had not a significant relationship with the patient's overall survival (OS) (17). Previous studies had shown that histology codes of PC were controversial in predicting cancer progression (18,19). Radiation therapy has significant effects on the management of penile squamous cell carcinoma, which enables sustained local control of the primary tumor while retaining functional anatomy (20). Therefore, we sought to develop a prognostic model incorporating include patient status, tumor characteristics and therapeutic methods based on large samples.
The nomograms are accurate and convenient clinical outcomes prediction tool, which are used to predict the prognosis of patients with malignancy (21,22). As far as we know, this method has been widely used in renal cell carcinoma, prostate cancer and bladder cancer (23-25). Our study aimed to establish nomograms based on the Surveillance, Epidemiology, and End Result (SEER) database to effectively evaluate and predict prognosis in PC patients. For this reason, clinicians can better counsel patients and tailor personalized treatment based on easily accessible clinical variables.
Methods
Data source and Study population
Patients diagnosed with PC from 2010 to 2015 were identified and extracted from SEER database using the SEER*Stat software (Version 8.3.6; National Cancer Institute, Bethesda, USA). The National Cancer Institute's SEER program is the largest population-based cancer database in the United States, which collects and publishes information of cancer patients in 18 registries, covering nearly 30% of the USA population (26). Patients included in our study should meet the following criteria: (I) diagnosed as PC (International Classification of Diseases for Oncology: 8,000/3, 8,010/3, 8,033/3, 8,051/3, 8,052/3, 8,070/3, 8,072/3, 8,073/3, 8,074/3, 8,076/3, 8,081/3, 8,083/3, 8,090/3, 8,092/3, 8,094/3, 8,097/3, 8,120/3, 8,140/3, 8,255/3, 8,403/3, 8,413/3, 8,480/3, 8,542/3, 8,940/3) with positive histology; (II) diagnosed from 2010 to 2015 to ensure a relatively long follow-up period; (III) complete data were available with active follow-up. Meanwhile, the exclusion criteria were as follows: (I) the first primary malignancy was not PC; (II) data on included variables were missing, such as race, age, sex, TNM stage, treatment methods and so on. The TNM stage were defined according to the 7th edition American Joint Committee on Cancer (AJCC) staging system (27). For each patient, we then collected the following information, including demographic characteristics (age race, and marital status), clinicopathological features (histology codes and TNM stage), use of surgery/lymph node removal (LNR)/radiation/chemotherapy, and survival outcomes (survival months, cancer-specific death, and cause of death). Primary endpoints of this research OS and cancer-specific survival (CSS). No medical ethics review was sought because of the identified data from public-use data.
Statistical analyses
To develop and validate the survival nomograms, patients were divided into two cohorts (the training cohort and the validation cohort) randomly at a ratio of 7:3 with the method of random-number generation (28,29). Comparisons of clinical information between two groups were made using the chi-square test. Uni- and multivariate Cox regression analysis were used to find the significant variables for OS and CSS. Based on the results of the multivariate analysis, nomograms models were constructed to predict 3-and 5-year OS and CSS. Furthermore, survival curves developed by Kaplan-Meier (KM) analysis and were compared utilizing the log-rank test among different variables.
Lastly, predictive performance of the nomograms was evaluated both internally (training set) and externally (validation set) with the calibration curve, Harrell's concordance index (C-index) (30) and the receiver operating characteristic (ROC) curve (31,32). Generally, area under the ROC curve (AUC) and the C-index range from 0.5 to 1.0, with 1.0 suggesting a perfect discrimination ability and 0.5 indicating the total chance (33). Consistency between the actual survival and the predicted survival was explored by calibration curves.
Chi-square test and Cox analysis were developed by SPSS 23.0 software (SPSS Inc, Chicago, IL, USA). Development and validation of the nomograms were performed using R version 3.6. 1 (http://www.r-project.org/) with rms, foreign, survival, survival ROC, and caret packages. During the whole analysis process, P<0.05 was considered to be statistically significant (two-sided).
Results
Demographic and characteristics of patients
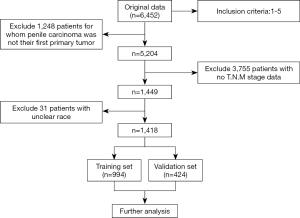
A total of 1,418 patients were enrolled in the study. A flow diagram of data selection was present in Figure 1. All eligible cases were randomly regrouped into training (n=994) and validation (n=424) cohorts. The demographic characteristics, clinicopathological features, and treatment methods of participants were shown in Table 1. No significant differences were detected between two cohorts in all variables (all P>0.05) except histology (P=0.038). Patients with squamous cell carcinoma (SCC) in the validation cohort were more than those in the training cohort significantly (P=0.038). We thought it was because of the relatively fewer patients of other types of PC patients, which led to the selection bias.
Table 1
| Total (n=1,418), n (%) | Training cohort (n=994), n (%) | Validation cohort (n=424), n (%) | P | |
|---|---|---|---|---|
| Age | 0.993 | |||
| <40 | 60 (4.3) | 42 (4.2) | 18 (4.2) | |
| 40–59 | 419 (29.5) | 292 (29.4) | 127 (30.0) | |
| 60–79 | 696 (49.1) | 488 (49.1) | 208 (49.1) | |
| ≥80 | 243 (17.1) | 172 (17.3) | 71 (16.7) | |
| Race | 0.739 | |||
| White | 1,187 (83.7) | 832 (83.7) | 355 (83.7) | |
| Black | 146 (10.3) | 105 (10.6) | 41 (9.7) | |
| Other | 85 (6.0) | 57 (5.7) | 28 (6.6) | |
| T stage | 0.529 | |||
| Ta | 19 (1.3) | 13 (1.3) | 6 (1.4) | |
| T1 | 805 (56.8) | 573 (57.7) | 232 (54.7) | |
| T2 | 320 (22.5) | 219 (22.0) | 101 (23.8) | |
| T3 | 232 (16.4) | 164 (16.5) | 68 (16.1) | |
| T4 | 42 (3.0) | 25 (2.5) | 17 (4.0) | |
| N stage | 0.726 | |||
| N0 | 1,128 (79.6) | 790 (79.5) | 338 (79.7) | |
| N1 | 84 (5.9) | 56 (5.6) | 28 (6.6) | |
| N2 | 98 (6.9) | 68 (6.9) | 30 (7.1) | |
| N3 | 108 (7.6) | 80 (8.0) | 28 (6.6) | |
| M stage | 0.796 | |||
| M0 | 1,364 (96.2) | 957 (96.3) | 407 (96.0) | |
| M1 | 54 (3.8) | 37 (3.7) | 17 (4.0) | |
| LNR | 0.349 | |||
| No/Biopsy only | 1,139 (80.3) | 792 (79.7) | 347 (81.8) | |
| Yes | 279 (19.7) | 202 (20.3) | 77 (18.2) | |
| Surgery | 0.841 | |||
| NO/unknown | 104 (7.3) | 72 (7.2) | 32 (7.5) | |
| Yes | 1,314 (92.7) | 922 (92.8) | 392 (92.5) | |
| Histology | 0.038 | |||
| Other | 128 (9.0) | 100 (10.1) | 28 (6.6) | |
| SCC | 1,290 (91.0) | 894 (89.9) | 396 (93.4) | |
| Radiation | 0.285 | |||
| No/Unknown | 1,299 (91.6) | 914 (92.0) | 385 (90.8) | |
| Yes | 119 (8.4) | 80 (8.0) | 39 (9.2) | |
| Chemotherapy | 0.582 | |||
| No/Unknown | 1,230 (86.7) | 859 (86.4) | 371 (87.5) | |
| Yes | 188 (13.3) | 135 (13.6) | 53 (12.5) | |
| Marital status | 0.623 | |||
| Married | 747 (52.7) | 530 (53.3) | 217 (51.2) | |
| Never married | 403 (28.4) | 275 (27.7) | 128 (30.2) | |
| Previously married | 268 (18.9) | 189 (19.0) | 79 (18.6) |
LNR, lymph nodes removal; SCC, Squamous cell carcinoma.
Cox regression analyses and KM curve analyses
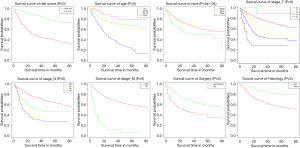
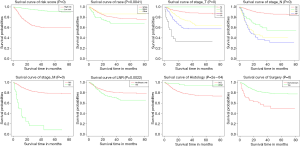
Uni- and multivariate Cox regression analyses were constructed to pick out key factors for OS and CSS. As shown in Table 2 and Table 3, 7 factors (including age, race, T stage, N stage, M stage, histology codes, and surgery, all P<0.05) were tightly associated with OS and 7 factors (including race, T stage, N stage, M stage, histology codes, surgery, and LNR, all P<0.05) were closely related to CSS. Subsequently, Finally, KM survival curves for OS and CSS were generated to learn the actual effect of different variables (Figures 2,3).
Table 2
| Characteristic | Univariate Cox | Multivariate Cox | |||||
|---|---|---|---|---|---|---|---|
| HR | P | 95% CI | HR | P | 95% CI | ||
| Age (year) | 0.000 | 0.000 | |||||
| <40 | REF | REF | |||||
| 40–59 | 1.96 | 0.066 | 0.957–4.017 | 1.774 | 0.181 | 0.767–4.106 | |
| 60–79 | 2.292 | 0.021 | 1.132–4.639 | 2.159 | 0.068 | 0.945–4.936 | |
| ≥80 | 5.802 | 0.000 | 2.849–11.818 | 7.08 | 0.000 | 3.07–16.331 | |
| Race | 0.003 | 0.000 | |||||
| White | REF | REF | |||||
| Black | 1.333 | 0.039 | 1.014–1.751 | 1.999 | 0.000 | 1.467–2.725 | |
| Others | 1.056 | 0.768 | 0.733–1.522 | 0.949 | 0.826 | 0.593–1.518 | |
| T stage | 0.000 | 0.000 | |||||
| T1 | REF | REF | |||||
| Ta | 0.29 | 0.217 | 0.04–2.07 | 0.603 | 0.625 | 0.079–4.586 | |
| T2 | 1.957 | 0.000 | 1.511–2.535 | 1.45 | 0.009 | 1.098–1.915 | |
| T3 | 2.716 | 0.000 | 2.08–3.548 | 2.059 | 0.000 | 1.535–2.762 | |
| T4 | 4.895 | 0.000 | 2.958–8.099 | 3.697 | 0.000 | 2.099–6.514 | |
| N stage | 0.000 | 0.000 | |||||
| N0 | REF | REF | |||||
| N1 | 2.167 | 0.000 | 1.571–2.988 | 1.741 | 0.011 | 1.134–2.674 | |
| N2 | 2.72 | 0.000 | 2.027–3.649 | 1.93 | 0.002 | 1.272–2.926 | |
| N3 | 3.607 | 0.000 | 2.79–4.665 | 2.267 | 0.000 | 1.528–3.364 | |
| M stage | 0.000 | 0.000 | |||||
| M0 | REF | REF | |||||
| M1 | 5.807 | 0.000 | 4.24–7.953 | 3.552 | 0.000 | 2.359–5.348 | |
| Histology | 0.000 | 0.012 | |||||
| SCC | REF | REF | |||||
| Others | 0.347 | 0.000 | 0.22–0.549 | 0.507 | 0.012 | 0.299–0.859 | |
| Surgery | 0.000 | 0.017 | |||||
| No/unknown | REF | REF | |||||
| Yes | 0.454 | 0.000 | 0.342–0.603 | 0.638 | 0.017 | 0.441–0.922 | |
| LNR | 0.350 | ||||||
| No/Biopsy only | REF | ||||||
| Yes | 1.043 | 0.707 | 0.837–1.299 | ||||
| Radiation | 0.001 | 0.449 | |||||
| No/unknown | REF | REF | |||||
| Yes | 1.689 | 0.000 | 1.287–2.217 | 0.868 | 0.446 | 0.604–1.248 | |
| Chemotherapy | 0.000 | 0.873 | |||||
| No/unknown | REF | REF | |||||
| Yes | 1.911 | 0.000 | 1.525–2.395 | 0.962 | 0.823 | 0.686–1.349 | |
| Marital status | 0.002 | 0.078 | |||||
| Married | REF | REF | |||||
| Previously married | 1.492 | 0.000 | 1.194–1.864 | 1.253 | 0.104 | 0.955–1.644 | |
| Never married | 1.288 | 0.022 | 1.037–1.599 | 1.309 | 0.042 | 1.01–1.698 | |
OS, overall survival; HR, Hazard ratio, CI, confidence interval; SCC, squamous cell carcinoma; LNR, lymph node removal; REF, reference.
Table 3
| Univariate Cox | Multivariate Cox | ||||||
|---|---|---|---|---|---|---|---|
| HR | P | 95% CI | HR | P | 95% CI | ||
| Age(year) | 0.131 | ||||||
| <40 | REF | ||||||
| 40–59 | 1.413 | 0.421 | 0.609–3.28 | ||||
| 60–79 | 1.223 | 0.633 | 0.535–2.799 | ||||
| ≥80 | 1.901 | 0.145 | 0.802–4.507 | ||||
| Race | 0.019 | 0.009 | |||||
| White | REF | REF | |||||
| Black | 1.695 | 0.01 | 1.132–2.538 | 1.806 | 0.005 | 1.192–2.738 | |
| Others | 0.68 | 0.319 | 0.319–1.452 | 0.617 | 0.219 | 0.285–1.334 | |
| T stage | 0.000 | 0.000 | |||||
| T1 | REF | REF | |||||
| Ta | 0 | 0.993 | 0–Inf | 0 | 0.993 | 0–Inf | |
| T2 | 2.867 | 0.000 | 1.982–4.147 | 1.942 | 0.001 | 1.306–2.888 | |
| T3 | 4.456 | 0.000 | 3.079–6.448 | 2.599 | 0.000 | 1.707–3.957 | |
| T4 | 11.706 | 0.000 | 6.711–20.417 | 4.732 | 0.000 | 2.475–9.047 | |
| N stage | 0.000 | 0.000 | |||||
| N0 | REF | REF | |||||
| N1 | 4.189 | 0.000 | 2.571–6.824 | 3.678 | 0.000 | 2.112–6.406 | |
| N2 | 7.085 | 0.000 | 4.748–10.573 | 4.371 | 0.000 | 2.61–7.32 | |
| N3 | 8.15 | 0.000 | 5.688–11.678 | 4.604 | 0.000 | 2.724–7.784 | |
| M stage | 0.000 | 0.000 | |||||
| M0 | REF | REF | |||||
| M1 | 10.013 | 0.000 | 6.613–15.163 | 3.091 | 0.000 | 1.927–4.957 | |
| Histology | 0.001 | 0.012 | |||||
| SCC | REF | REF | |||||
| Others | 0.212 | 0.001 | 0.087–0.515 | 0.308 | 0.012 | 0.124–0.768 | |
| Surgery | 0.000 | 0.028 | |||||
| No/unknown | REF | REF | |||||
| Yes | 0.32 | 0.000 | 0.214–0.479 | 0.603 | 0.029 | 0.384–0.949 | |
| LNR | 0.002 | 0.031 | |||||
| No/Biopsy only | REF | REF | |||||
| Yes | 1.635 | 0.002 | 1.191–2.244 | 0.653 | 0.030 | 0.444–0.959 | |
| Radiation | 0.000 | 0.619 | |||||
| No/unknown | REF | REF | |||||
| Yes | 2.51 | 0.000 | 1.697–3.712 | 0.892 | 0.608 | 0.576–1.381 | |
| Chemotherapy | 0.000 | 0.765 | |||||
| No/unknown | REF | REF | |||||
| Yes | 3.51 | 0.000 | 2.573–4.787 | 0.936 | 0.75 | 0.625–1.403 | |
| Marital status | 0.033 | 0.158 | |||||
| Married | REF | REF | |||||
| Previously married | 1.573 | 0.014 | 1.096–2.256 | 1.442 | 0.055 | 0.992–2.095 | |
| Never married | 1.348 | 0.086 | 0.959–1.895 | 1.18 | 0.358 | 0.829–1.679 | |
CSS, cancer-specific survival; HR, hazard ratio, CI, confidence interval; SCC, squamous cell carcinoma; LNR, lymph node removal; REF, reference.
Nomograms construction and validation
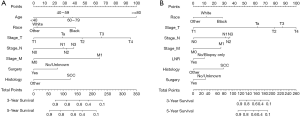
The nomograms were established for predicting OS and CSS, the OS (Figure 4A) nomogram revealed that age had the most important contributions to prognosis, followed by the T stage, M stage, race, N stage, histology codes, and surgery. As for CSS (Figure 4B), the nomogram demonstrated that the T stage contributed most to prognosis, followed by N stage, histology codes, race, M stage, surgery, and LNR.
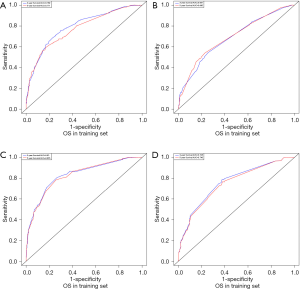
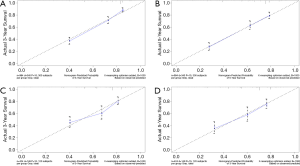
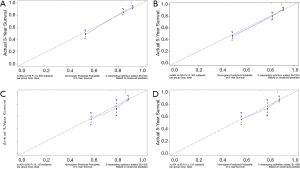
C-index, calibration curve, ROC curve, and AUC were used to validate the accuracy of nomograms internally (training cohort) and externally (validation cohort). The C-indices for OS and CSS were 0.755 and 0.805 in the training cohort and 0.711 and 0.737 in the validation cohort. In addition, the 3- and 5-year AUCs for OS were 0.792 and 0.771 in the training cohort (Figure 5A), and 0.687 and 0.695 in the validation group (Figure 5B). When it came to CSS, it was 0.83 and 0.826 in the training cohort (Figure 5C) and 0.758 and 0.746 in the validation cohort (Figure 5D). Additionally, there were superb consistency between the calibration curves and the 45-degree reference lines in the calibration plots of 3 and 5-year OS (Figure 6) and CSS (Figure 7), which suggested that calibration curves for nomograms predicted 3 and 5-year OS and CSS performed pretty well with the ideal model.
Discussion
Nomograms are widely used to predict cancer survival because of their intuitive presentation of data, accuracy, and personalization (21). Combining different prognostic factors, we successfully developed and preliminarily validated nomograms to forecast 3- and 5-year OS and CSS of PC patients. The nomograms revealed favorable discrimination and good calibration both in internal and external validations. As a result, the nomogram of OS incorporated seven factors, including age at diagnosis, race, TNM stage, and histology codes, while the nomogram of CSS including seven factors, race, TNM stage, LNR, histology codes, and surgical treatment.
Previous studies have identified several risk factors to be independent prognostic factors for patients with PC (11-13,34-37). However, these studies focused on limited key factors but ignored some other significant risk factors and the sample of these was limited. In order to better predict prognosis for PC patients, we constructed a more synthetical model based on a large number of samples of 1,418 cases.
On the basis of the KM methods and log-rank analyses, survival rates were self-evidently affected by age and race. Of the eligible patients, it had been shown that getting older had a direct impact on OS. Previous research has suggested that marital status had protection on the OS and CSS of PC (15,29,34). However, our research found that marital status was not a significant predictor, probably because this research included only those patients who had poor physical conditions and a higher degree of cancer risk (15). Some studies also have examined the relationship between different races. Rippentrop et al. (38) found that notable differences could be detected in survival between African Americans and whites. Sharma et al. (36) described that being black was related to worse OS. Our study confirmed that the black race was associated with lower OS and CSS than white, which coincided with the former report.
As we all know, tumor characteristics play an important role in the prognosis of cancer patients. We established the first practical nomograms illustrated that SCC had a negative impact on patient survival than other pathological types, perhaps because patients with other pathological types of tumor which chosen more aggressive treatment, this finding needs further study to confirm. Our results shown that more advanced T, N and M stages meant potential predictors of PC patients. Based on the established nomograms, we could know that the T and N stages were great significant prognostic factors. The N stage was noticeably associated with OS and CSS according to our analysis, previous research found that lymph node dissection was one of the most important prognostic factors in men with PC (39-41), also, early lymph node dissection is recommend by many guidelines (35,42).
Previous studies have shown that LNR was positively associated with CSS (36,37), which was also confirmed by our study. However, OS of patients were not reelevate to the LNR in our study. This might be related to the incorrect clinical examination of the nodal staging (43,44), or the lymphadenectomy complication rate was relatively high. Hakenberg et al. (35) confirmed that surgical resection represented a prognostic factor for survival, which was consistent with our results. Furthermore, chemotherapy and radiotherapy were not independently correlated with OS or CSS in our study. Previous studies also suggested that chemotherapy was frustratingly ineffective in PC (9). In addition, the 2014 European Association of Urology (EAU) guidelines did not advocated radiation because the results of patients who received radiotherapy were discouraging (35). A lack of detailed information about chemotherapy and radiotherapy in the database might also lead to an inability to accurately identify prognostic factors. Interestingly, recent study established a competing-risks analysis model for PC patients (45), however, our research seems to be more suitable for the practical needs of clinical work, because our nomograms could visually and make individualized predictions.
Although these two nomograms performed well, we acknowledged that our study had some shortcomings. First of all, it was important to note that only a third of the patients were enrolled in our study, we excluded other patients because of the lack of necessary clinical data. Second, the nomograms lacked of some key indicators which were missing in the SEER database, such as biological markers, genetic mutation, site of involvement, and specific types of surgery. Furthermore, since all patients included in our study were from the same SEER dataset, our nomograms cannot be verified from another database.
Conclusions
It was the first time to conduct survival models for PC patients with predictive performance. It might be valuable of clinical application and further exploration with more studies in the future.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr.2020.03.77). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Barnholtz-Sloan JS, Maldonado JL, Pow-sang J, et al. Incidence trends in primary malignant penile cancer. Urol Oncol 2007;25:361-7. [Crossref] [PubMed]
- Bray F, Klint A, Gislum M, et al. Trends in survival of patients diagnosed with male genital cancers in the Nordic countries 1964-2003 followed up until the end of 2006. Acta Oncol 2010;49:644-54. [Crossref] [PubMed]
- Backes DM, Kurman RJ, Pimenta JM, et al. Systematic review of human papillomavirus prevalence in invasive penile cancer. Cancer Causes Control 2009;20:449-57. [Crossref] [PubMed]
- Coelho RWP, Pinho JD, Moreno JS, et al. Penile cancer in Maranhao, Northeast Brazil: the highest incidence globally? BMC Urol 2018;18:50. [Crossref] [PubMed]
- Shabbir M, Kayes O, Minhas S. Challenges and controversies in the management of penile cancer. Nat Rev Urol 2014;11:702-11. [Crossref] [PubMed]
- Bevan-Thomas R, Slaton JW, Pettaway CA. Contemporary morbidity from lymphadenectomy for penile squamous cell carcinoma: the M.D. Anderson Cancer Center Experience. J Urol 2002;167:1638-42. [Crossref] [PubMed]
- Kroon BK, Horenblas S, Lont AP, et al. Patients with penile carcinoma benefit from immediate resection of clinically occult lymph node metastases. J Urol 2005;173:816-9. [Crossref] [PubMed]
- Necchi A, Pond GR, Raggi D, et al. Clinical Outcomes of Perioperative Chemotherapy in Patients With Locally Advanced Penile Squamous-Cell Carcinoma: Results of a Multicenter Analysis. Clin Genitourin Cancer 2017;15:548-55.e3. [Crossref] [PubMed]
- Robinson R, Marconi L, MacPepple E, et al. Risks and Benefits of Adjuvant Radiotherapy After Inguinal Lymphadenectomy in Node-positive Penile Cancer: A Systematic Review by the European Association of Urology Penile Cancer Guidelines Panel. Eur Urol 2018;74:76-83. [Crossref] [PubMed]
- Thuret R, Sun M, Abdollah F, et al. Tumor grade improves the prognostic ability of American Joint Committee on Cancer stage in patients with penile carcinoma. J Urol 2011;185:501-7. [Crossref] [PubMed]
- Emerson RE, Ulbright TM, Eble JN, et al. Predicting cancer progression in patients with penile squamous cell carcinoma: the importance of depth of invasion and vascular invasion. Mod Pathol 2001;14:963-8. [Crossref] [PubMed]
- Zhou X, Qi F, Zhou R, et al. The role of perineural invasion in penile cancer: a meta-analysis and systematic review. Biosci Rep 2018; [Crossref] [PubMed]
- Ficarra V, Zattoni F, Cunico SC, et al. Lymphatic and vascular embolizations are independent predictive variables of inguinal lymph node involvement in patients with squamous cell carcinoma of the penis: Gruppo Uro-Oncologico del Nord Est (Northeast Uro-Oncological Group) Penile Cancer data base data. Cancer 2005;103:2507-16. [Crossref] [PubMed]
- Mao W, Zhang Z, Huang X, et al. Marital Status and Survival in Patients with Penile Cancer. J Cancer 2019;10:2661-9. [Crossref] [PubMed]
- Cubilla AL. The role of pathologic prognostic factors in squamous cell carcinoma of the penis. World J Urol 2009;27:169-77. [Crossref] [PubMed]
- McDougal WS. Carcinoma of the penis: improved survival by early regional lymphadenectomy based on the histological grade and depth of invasion of the primary lesion. J Urol 1995;154:1364-6. [Crossref] [PubMed]
- Cubilla AL, Reuter V, Velazquez E, et al. Histologic classification of penile carcinoma and its relation to outcome in 61 patients with primary resection. Int J Surg Pathol 2001;9:111-20. [Crossref] [PubMed]
- Renaud-Vilmer C, Cavelier-Balloy B, Verola O, et al. Analysis of alterations adjacent to invasive squamous cell carcinoma of the penis and their relationship with associated carcinoma. J Am Acad Dermatol 2010;62:284-90. [Crossref] [PubMed]
- Diorio GJ, Leone AR, Spiess PE. Management of Penile Cancer. Urology 2016;96:15-21. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Balachandran VP, Gonen M, Smith JJ, et al. Nomograms in oncology: more than meets the eye. Lancet Oncol 2015;16:e173-80. [Crossref] [PubMed]
- Jiang T, Wu YP, Chen SH, et al. Prognosis and clinicopathological characteristics of renal cell carcinoma: does bilateral occurrence influence overall and cancer-specific survival? Transl Cancer Res 2020;9:432-40. [Crossref]
- Bandini M, Marchioni M, Pompe RS, et al. First North American validation and head-to-head comparison of four preoperative nomograms for prediction of lymph node invasion before radical prostatectomy. BJU Int 2018;121:592-9. [Crossref] [PubMed]
- Zhang W, Wang R, Ma W, et al. Systemic immune-inflammation index predicts prognosis of bladder cancer patients after radical cystectomy. Ann Transl Med 2019;7:431. [Crossref] [PubMed]
- Cronin KA, Ries LA, Edwards BK. The Surveillance, Epidemiology, and End Results (SEER) Program of the National Cancer Institute. Cancer 2014;120:3755-7. [Crossref] [PubMed]
- Leijte JA, Gallee M, Antonini N, et al. Evaluation of current TNM classification of penile carcinoma. J Urol 2008;180:933-8; discussion 8. [Crossref] [PubMed]
- Collins GS, Reitsma JB, Altman DG, et al. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): The TRIPOD Statement. Eur Urol 2015;67:1142-51. [Crossref] [PubMed]
- Lei Z, Li J, Wu D, et al. Nomogram for Preoperative Estimation of Microvascular Invasion Risk in Hepatitis B Virus-Related Hepatocellular Carcinoma Within the Milan Criteria. JAMA Surg 2016;151:356-63. [Crossref] [PubMed]
- Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361-87. [Crossref] [PubMed]
- Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982;143:29-36. [Crossref] [PubMed]
- Deng H, Qi X, Zhang Y, et al. Diagnostic accuracy of contrast-enhanced computed tomography for esophageal varices in liver cirrhosis: a retrospective observational study. J Evid Based Med 2017;10:46-52. [Crossref] [PubMed]
- Valentini V, van Stiphout RG, Lammering G, et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol 2011;29:3163-72. [Crossref] [PubMed]
- Thuret R, Sun M, Budaus L, et al. A population-based analysis of the effect of marital status on overall and cancer-specific mortality in patients with squamous cell carcinoma of the penis. Cancer Causes Control 2013;24:71-9. [Crossref] [PubMed]
- Hakenberg OW, Comperat EM, Minhas S, et al. EAU guidelines on penile cancer: 2014 update. Eur Urol 2015;67:142-50. [Crossref] [PubMed]
- Sharma P, Spiess PE. Penile cancer: Advanced penile cancer continues to elude systemic therapy. Nat Rev Urol 2015;12:185-6. [Crossref] [PubMed]
- Leone A, Diorio GJ, Pettaway C, et al. Contemporary management of patients with penile cancer and lymph node metastasis. Nat Rev Urol 2017;14:335-47. [Crossref] [PubMed]
- Rippentrop JM, Joslyn SA, Konety BR. Squamous cell carcinoma of the penis: evaluation of data from the surveillance, epidemiology, and end results program. Cancer 2004;101:1357-63. [Crossref] [PubMed]
- Li Z, Guo S, Wu Z, et al. Proposal for reclassification of N staging system in penile cancer patients, based on number of positive lymph nodes. Cancer Sci 2018;109:764-70. [Crossref] [PubMed]
- Soodana-Prakash N, Koru-Sengul T, Miao F, et al. Lymph node yield as a predictor of overall survival following inguinal lymphadenectomy for penile cancer. Urol Oncol 2018;36:471.e19-471.e27. [Crossref] [PubMed]
- Mao W, Huang X, Kong M, et al. More lymph node dissection improves survival in patients with newly diagnosed lymph node-positive penile cancer. Int Urol Nephrol 2019;51:641-54. [Crossref] [PubMed]
- Clark PE, Spiess PE, Agarwal N, et al. Penile cancer: Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2013;11:594-615. [Crossref] [PubMed]
- Heyns CF, Fleshner N, Sangar V, et al. Management of the lymph nodes in penile cancer. Urology 2010;76:S43-57. [Crossref] [PubMed]
- Pizzocaro G, Algaba F, Horenblas S, et al. EAU penile cancer guidelines 2009. Eur Urol 2010;57:1002-12. [Crossref] [PubMed]
- Yang J, Pan Z, He Y, et al. Competing-risks model for predicting the prognosis of penile cancer based on the SEER database. Cancer Med 2019;8:7881-9. [Crossref] [PubMed]