
Differential expression of microRNAs in xenografted Lewis lung carcinomas subjected to intermittent hypoxia: a next-generation sequence analysis
Introduction
Obstructive sleep apnea (OSA) has a high prevalence among middle-age population (1). The recurrent collapse of upper airway during sleep contributes to intermittent hypoxia (IH) and sleep fragmentation (2), two of the novel pathophysiological features of OSA patients. Robust evidence has confirmed the association between OSA, cardiovascular, cognitive and metabolic abnormalities (2). Recent, epidemiological investigations elucidated that the incidence and mortality of tumor in subjects with OSA were higher than those without OSA. The polysomnographic variables, apnea-hypopnea index, and oxygen desaturation index, are closely associated with tumor progression (3-5). Focusing on xenografting with Lewi lung cancer cells, epithelial tumor cells TC1 and melanoma cells, multiple experimental studies, that included ours, proved that IH is closely correlated with tumor growth, invasion, and metastasis (6-9). The potential mechanisms of how IH accelerates tumor progression, however, are still far from conclusion.
MicroRNAs (MiRNAs), the non-coding small RNAs with the length of 22 nucleotides, are a group of newly discovered endogenous single-strand RNAs (10). MiRNAs regulate gene expression through binding to the target mRNAs at 3’ untranslated regions of mRNAs to inhibit translation or to promote mRNA degradation (11). Previous studies indicated that miRNAs might present as tumor suppressors or oncogenes, inhibiting or promoting tumor progression and angiogenesis (12). Furthermore, miRNAs were considered as biomarkers of tumor diagnosis and prognosis, and even as the anti-tumor therapeutic target (13).
Hypoxia is a reduction of tissue oxygen tension. Tissue hypoxia influences various cell functions. Previous study (14) demonstrated that change of several miRNAs was detected in cells exposed to hypoxia, which could promote tumor progression. Several hypoxia-induced miRNAs, such as miRNA-210, play a novel role in the hypoxic adaptation of cancer cells (15,16), in which hypoxia-inducible factor-1α (HIF-1α) performs a vital role in hypoxia-induced miRNA changes.
Several previous studies paid attention to the miRNA profile among OSA patients or IH animal models. A study from Spain (17) elucidated that circulating miRNA profile could be as a potential biomarker for the diagnosis of OSA. Gao and colleagues (18) utilizing miRNA microarray in an IH-induced rat model proved that some miRNAs could be involved in the OSA-induced cognitive impairments. Furthermore, a part of studies addressed the miRNAs alteration in OSA patients with cancer. Almendros and co-workers (19) studied the effect of circulating exosomes from OSA patients on tumor malignant properties. Their results showed that exosomes from OSA patients enhanced the properties of lung tumor cell and eleven distinct miRNAs were observed in IH-exposed mice. Freitas et al. (20) found that severe OSA is independently associated with increased miRNAs (miR-254 and miR-320e) which involved in various cancers.
Although some researches already addressed the relationship between miRNAs, tumor and IH, only microarray analysis and real-time polymerase chain reaction (RT-PCR) were conducted in those studies. Therefore, the detectable numbers of differential expressed miRNAs were limited and large-scale screen were failed to perform. The purpose of our study is to screen the aberrant miRNAs expressions in Lewis lung carcinoma (LLC) xenograft mice exposed to IH via the new next-generation sequence (NGS) method and to classify the influence of IH on tumor miRNAs. We present the following article in accordance with the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-2913.
Methods
Animal grouping
This protocol was approved by the IACUC and IBC Committee in Zhongshan Hospital, Xiamen University (approved number: 2017-015) with a tumor volume less than 5,000 mm3, and conducted in accordance with the Guide for the Care and Use of Laboratory Animal. Twenty four 7-week-old male C57BL/6J mice (6,19,21-23) were purchased from the Chinese Academy of Science Laboratory Animals Center in Shanghai, China, and were randomly assigned to either the normoxia (control, CTL) group or the IH group (n=12 in each group). Except the IH period, all mice were housed with standard cages in a 12:12 hour light-dark cycle environment, were fed and had tap water ad libitum. Mice were weighed each week. The general condition of mice, such as appearance, weight loss, ruffled fur, reduced mobility, and body posture, were observed daily, and no human endpoints were used. If the tumor volume was ≥5,000 mm3, euthanization was carried out immediately. No mice were euthanized prematurely, and all mice underwent euthanasia through anesthetized with pentobarbital at the experimental endpoint.
Cells culture
LLC cells were obtained from CoBioer Biosciences Co., Ltd. (Shanghai, China), and were cultured according to the protocol of the manufacturer and our previous studies (8,23).
The IH exposure
The IH protocol was the same as our previous studies (8,23,24), with some modification. Mice in the IH group were exposed in a self-made plexiglass chamber with one-way valves. Three types of gases: nitrogen, oxygen, and compressed air, flowed into the chamber and were controlled by a programmable instrument. The oxygen concentration in the chamber fluctuated between normal (21%±1%) to nadir (6%±1%) for every 120 seconds of the IH cycle (from hypoxia to reoxygenation): 50 s of nitrogen, resting for 10 s, oxygen for 10 s, and compressed air for 50 s. The IH exposure lasted for 8 hours per day, from 9 AM to 5 PM in the light-on period. The experimental period was 5 weeks (9,21,22,25,26).
Subcutaneous lung tumor induction and assessment
In order to create a lung carcinoma xenograft model in mice (21,26,27), the following procedure was done: after one week of the IH exposure, the LLC cells (1×106 LLC/100 µL PBS per mouse) were injected into the right flank of each mouse. Once a tumor became palpable, the length (L) and width (W) of tumor in mice were measured every 5 days, and the tumor volume (mm3) was calculated as W2×L/2.
Tissue preparation
After 5 weeks of the experiment, all mice were deeply anesthetized with pentobarbital. Tumors were excised and weighted. Tumor tissues (0.5 cm3) were stored in RNA locker (Beijing Tiandz, Inc. China), and then were transferred to −80 °C refrigerator for further analysis. In this study, three tumor tissue samples from each group were used to perform NGS (20,28).
MiRNA expression profile analysis using NGS technology
MiRNAs were sequenced by the BGISEQ-500 as previously described (28,29). MiRNA sequencing and bioinformatics analysis were performed in the Beijing Genomics Institute of BGI-Shenzhen company (Shenzhen, China).
RNA extraction
Total RNA was extracted from the tumor tissue using Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacture's instruction. Grind about 60 mg with liquid nitrogen into powder and the powder samples were transferred into the 2 mL tube with 1.5 mL Trizol reagent. The mix was further centrifuge at 12,000 ×g for 5 min at 4 °C. The supernatant was transferred to a new 2.0 mL tube which was added with 0.3 mL of chloroform/isoamyl/alcohol (24:1) per 1.5 mL of Trizol reagent. After centrifugation, the aqueous phase was transferred to a new tube in which equal volume of supernatant of isopropyl alcohol was added. The mix was centrifuged, and then the supernatant was removed. After washed with 1 mL 75% ethanol, the RNA pellet was air-dried in the biosafety cabinet and then dissolved by add 25–100 µL of DEPC water. Subsequently, total RNA was qualified and quantified using Nano Drop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA).
MiRNA library construction
Library was prepared with 1 µg total RNA for each sample. Total RNA was purified by electrophoretic separation on a 15% urea denaturing polyacrylamide gel electrophoresis (PAGE) gel and small RNA regions corresponding to the 18–30 nt bands in the marker lane (14–30 ssRNA Ladder Marker, TAKARA) were excised and recovered. Then the 18–30 nt small RNAs were ligated to a 5'-adaptor and a 3'-adaptor. The adaptor-ligated small RNAs were then transcribed into cDNA with SuperScript II Reverse Transcriptase (Invitrogen, USA) and several rounds of PCR amplification with PCR Primer Cocktail and PCR Mix were conducted to enrich the cDNA fragments. The PCR products were selected by agarose gel electrophoresis with target fragments 100–120 bp, and then purified by QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA, USA). The library was qualified and quantitated in two methods: checking the distribution of the fragments size using the Agilent 2100 bioanalyzer, and quantifying the library using RT-PCR (TaqMan Probe). The final ligation PCR products were sequenced using the BGISEQ-500 platform (BGI-Shenzhen, China).
Prediction of novel miRNAs
The 49 nt sequence tags from sequencing would go through the data cleaning analysis first, which included getting rid of the low quality tags, 5' adaptor contaminants from the 50 nt tags, to get credible clean tags. Then the length distribution of the clean tags and common and specific sequences between samples would be summarized. Furthermore, the standard analysis would annotate the clean tags into different categories and took those which could not be annotated to any category to predict the novel miRNA. The novel miRNA should not be described in miRBase before.
Target gene prediction, gene ontology (GO), and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis
Two softwares were used to predict miRNA targets: miRanda (http://www.microrna.org/microrna/home.do) and TargetScan (http://www.targetscan.org/vert_71/). The target gene prediction was accessed by whether miRNAs maintained a regulatory role via the 3’ untranslated region of target genes. Only the genes identified simultaneously by both of the programs were considered the potential target genes. GO enrichment analysis (http://www.geneontology.org) was conducted to analyze the function of differentially expressed miRNAs. KEGG (http://www.genome.jp/kegg) was used to identify the main biochemical and signaling pathways. Only KEGG pathways with P value <0.05 and a false discovery rate <0.05 were retained (30).
Verification of the miRNA expression profile analysis data using qRT-PCR
RNAs from the tumors were extracted using miRNA isolation kit (Invitrogen) and were reversely transcribed using Taqman MicroRNA Reverse Transcription Kit (Life Technologies, Carlsbad, California, USA) following to the protocol of the manufacturer. U6 was used as the internal control for miRNA (31). The mature miRNA primers sequences were obtained from the miRBase. Quantitative PCR (qRT-PCR) was conducted with the ABI PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA, USA). All the primers used in this study are listed at http://fp.amegroups.cn/cms/b450465b00cb1e3c3349307f02a18fdf/TCR-19-2913-1.pdf (Designed by Sangon Biotech Co., Ltd., Shanghai, China). The efficiency of each primer was calculated using a standard curve (data not shown). All reactions were run in triplicate, and the data were analyzed by the 2−ΔΔCT method.
Statistical analysis
Data are expressed as mean ± standard deviation and was performed with GraphPad Prism software 5.0 (GraphPad Software, Inc., La Jolla, CA, USA). The non-parametric Wilcoxon signed-rank test was conducted to evaluate the differential expression between the CTL and IH groups. A P value of less than 0.05 was regarded as a significant difference.
Results
Bodyweight, tumor volume, and tumor weight
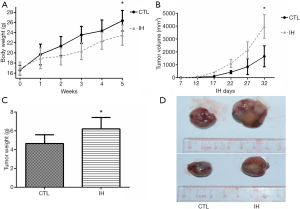
After 5 weeks of the experiment, none mouse died in each group, all mice were enrolled into analysis. Mice exposed to IH gained less body weight. Both tumor volume and weight were significantly higher in the IH group than those of the CTL group (Figure 1).
Differential expression of miRNAs and prediction of novel miRNAs
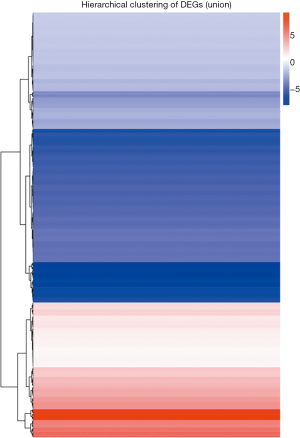
A total of 485 differentially expressed miRNAs were filtered (259 novel miRNAs and 226 known miRNAs). Of them, 154 miRNAs were upregulated and 331 miRNAs were downregulated (Figure 2 and see at http://fp.amegroups.cn/cms/dd60594e9aa9e00b113afece372875e3/TCR-19-2913-2.pdf). The log2 fold change values of top 10 miRNAs (5 known miRNAs and 5 novel miRNAs) are listed in Table 1. The results of cluster analysis are outlined in Figure 3.
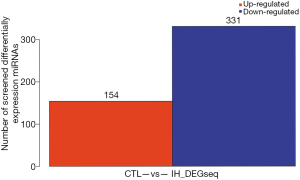
Table 1
| miRNA | Fold change, log2 ratio (IH/CTL) |
|---|---|
| miR-767 | 8.006 |
| miR-466f-5p | −7.413 |
| miR-5122 | −6.551 |
| miR-124-3p | 6.006 |
| miR-590-3p | 5.525 |
| novel-miR-140 | 9.298 |
| novel-miR-130 | 9.226 |
| novel-miR-301 | 9.028 |
| novel-miR-177 | 9.017 |
| novel-miR-90 | 8.835 |
CTL, normoxia, control; IH, intermittent hypoxia.
Predicted target genes, GO enrichment and KEGG pathway analysis
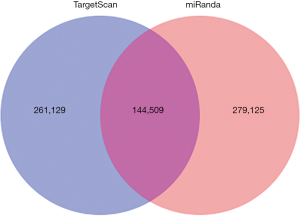
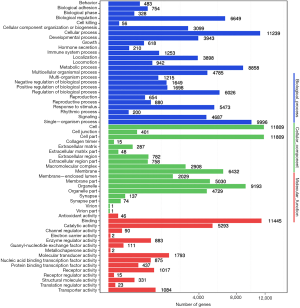
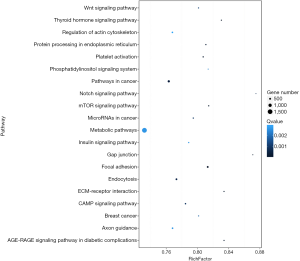
The predicted genes, GO enrichment, and KEGG pathway analysis of the abovementioned miRNAs are outlined at http://fp.amegroups.cn/cms/1b41d3aa5bb338fee148fd41c399bf2a/TCR-19-2913-3.pdf. Figure 4 shows the predicted target genes both in TargetScan and miRanda. In order to identify the biological functions of the target genes, GO and KEGG pathway analysis were conducted. A total of 59 significantly enriched GO terms were found, including 25 biological processes, 18 cell metabolic and biosynthetic processes, and 16 molecular functions. Each term contained various target genes (range from 1–1,1445 genes, the detail target gene numbers showed at the top of each bar in Figure 5). Meanwhile, 20 significantly KEGG pathways enrichment were detected, especially in metabolic, cancer, and focal adhesion pathways (Figure 6).
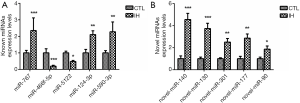
Validation of differentially expressed miRNAs by qRT-PCR
The top 5 discrepant miRNAs both in known and novel miRNAs were selected for further qRT-PCR analysis (31-33). Compared with the CTL group, miR-767, miR-124-3p and miR-590-3p were increased whereas miR-466f-5p and miR-5122 were decreased in the IH group. All novel miRNAs (miR-140, miR-130, miR-301, miR-177 and miR-90) demonstrated a significant upregulation in the IH group (Figure 7). The results of qRT-PCR were in concordance with the NGS data.
Discussion
In this study, the NGS analysis of miRNAs expression profiles elucidated several aberrantly expressed miRNAs in xenograft mice exposed to IH, an essential hallmark of OSA. The top 10 differentially expressed miRNAs were validated by qRT-PCR. Furthermore, bioinformatics results showed that those miRNAs regulated target genes were markedly enriched in abundant biological processes and pathways, such as biological processes, cell metabolic and biosynthetic processes and molecular functions.
An increasing number of studies indicated the close association between OSA and tumor incidence and mortality (3,34-36). The exactly molecular mechanism is still unclear. As a pathophysiological feature of OSA, IH plays an essential role in tumor progression. Previous in vivo studies (6-9,21,23,25) inferred that IH facilitated tumor progression, invasion, and metastasis.
MiRNAs are a class of small non-coding RNA molecules that regulate gene expression post-transcriptionally and influence several physiological processes (10). Their dysregulation is related to oncogenesis or tumor-suppression (13,37). Hypoxia, especially in the tumor microenvironment, regulates cellular progress, such as growth, metabolism, differentiation, and apoptosis. Hypoxia is also an important regulator of miRNAs biogenesis and function (38). The widespread effects of hypoxia on genes expression and cellular functions might be potentially mediated by some specific miRNAs (38,39). Hypoxia might influence tumor pathophysiological characteristics via the mediation of miRNAs (39).
Chronic hypoxia is divided into chronic sustained hypoxia and chronic IH. The pathophysiological features of the two types of hypoxia are discrepant. IH which is familiar to ischemia-reperfusion injury (40), easily produces oxidative stress and systemic inflammation (41,42). Most of the previous studies focused on the effect of chronic sustained hypoxia on miRNAs in the tumor (16,43,44). This study aimed to determine the aberrant miRNA profiles by NGS in a mouse model with tumor and IH. We conducted bioinformatics tools (GO and KEGG) to predict the target genes of those differentially expressed miRNAs and potential signaling pathways. The results of this study elucidated that a total of 485 differentially expressed miRNAs were filtered (259 novel miRNAs and 226 known miRNAs). Of them, 154 miRNAs were upregulated and 331 miRNAs were downregulated when tumor-bearing mice were exposed to IH. After bioinformatics analysis, numerous potential target genes and relevant signaling pathways, including biological process, cellular metabolism and molecular function were observed. The qRT-PCR was conducted to verify the top 10 differentially expressed miRNAs (top 5 known miRNAs and top 5 novel miRNAs, see at http://fp.amegroups.cn/cms/149cc371edaafb612c5ebc7f59179133/TCR-19-2913-4.pdf, http://fp.amegroups.cn/cms/4eb43d6421a2fe8717baad1f47542729/TCR-19-2913-5.pdf). Results confirmed the NGS data by showing that all novel miRNAs (miR-140, miR-130, miR-301, miR-177, and miR-90) were upregulated in mice exposed to IH; meanwhile, some known miRNAs (miR-767, miR-124-3p, and miR-590-3p) were upregulated and the others (miR-466f-5p and miR-5122) were downregulated. MiR-767 was found to promote cell proliferation in human melanoma (45). Previous studies showed that decreased miR-124-3p facilitates cancer growth and metastasis in solid tumors of different organs (46-48). Hypoxia appears to be closely associated with different expression of miRNAs in various diseases, especially in cancers. Evidence revealed that the expression of exosomal miRNAs in hypoxic esophageal squamous carcinoma cells was altered (44). MiR-140-5p enhanced hypoxia-induced cell injury partially via up-regulation of MLK3 in vitro (43). However, Hou et al. (49) showed that miR-140 inhibition increased vascular endothelial growth factor-A (VEGF-A) expression and promoted angiogenesis of hepatocellular carcinoma. In the present study, the miR-124-3p and miR-40 levels were increased in IH-induced mice. Whether the specific increased miRNAs (miR-124-3p and miR-140) induced by IH is a protective mechanism remains unrevealed. The discrepancy between studies and underlying molecular mechanism are required for further investigations. Regarding miR-590-3p, studies (50,51) also showed that the increased miR-590-3p could promote colon cancer cell proliferation and metastasis via Hippo or Wnt/beta-catenin signaling pathways. MiR-590-3p was also found to facilitate cervical and ovarian cancers as well (52,53). Hypoxia-induced miR-590-5p promotes colorectal cancer progression via modulating matrix metalloproteinase activity (54). Our study is consistent with previous studies by showing an increased IH-induced miR-590-3p expression. However, an in vitro study by Zhang et al. (55) reported that miR-590-3p significantly suppressed the expression of VEGF and reduced the activation of Wnt/beta-catenin signaling pathway. In addition, Bao et al. (56) showed that miR-590-3p prevented oxidized LDL-induced angiogenesis inhibiting oxidized LDL-induced upregulation of VEGF. These two studies (55,56) appeared to be controversial to one of our previous studies in which increased VEGF was observed in the tumor of IH-induced mice (23). This discrepancy might explain that our previous study addressed the effect of IH on VEGF in a xenografted tumor-bearing mice, while the study by Zhang et al. (55) addressed the effect of miR-590-3p on spiral artery remodeling in a trophoblast cell line, and the study by Bao et al. (56) paid attention to the effect of LINC00657 and miR-590-3p on the angiogenesis in a vascular endothelial cells. The present study demonstrated abundant potential target genes mediated by aberrantly expressed miRNAs after IH exposure. A large amount of differentially expressed miRNAs in IH-induce mice were screened, but the potential target genes mediating by miRNAs and signal pathways between IH, miRNAs and tumor were not yet included in our study. Further studies are needed to evaluate the exact role of certain miRNA in the pathogenesis of IH accelerating tumor progression.
Several limitations should be mentioned in this study. First, we detected the miRNAs in the whole tumor and it was difficult to distinguish whether the miRNAs came from intra or extracellular compartments or even bloodborne. Second, the function and relevance of identified differentially expressed miRNAs were not explored and the underlying signaling pathways between IH, aberrant miRNAs, and tumor progression were not investigated in this study. We speculated that some of the aberrant expression of miRNAs under IH condition served as suppressor. Additional studies are required to confirm whether certain differentially expressed miRNA correlates with IH-induced tumor progression through evaluating predicted target genes and potential signaling pathways. Third, IH which is different from constant hypoxia from the pathophysiology perspective (57,58), is more like ischemia-reperfusion injury (40). A constant hypoxia (22) group was not set as control group in this study, which might probably weaken our conclusions. Forth, female mice were not included in this study, hence the influence of gender on tumor biological behaviors could not be ruled out. Fifth, we only created a LLC xenograft mouse model and these results are possibly not transferred to other types of cancer.
Conclusions
This study found an abundance of differentially expressed miRNAs in xenograft mice exposed to IH. We postulated that those miRNAs might play a pivotal role in the mechanism of OSA aggravating tumor progression. Further studies are warranted to confirm the mediated effect of certain miRNA on predicted target genes and through specific signaling pathway.
Acknowledgments
We appreciated the help from the Beijing Genomics Institute of BGI-Shenzhen Company (Shenzhen, China).
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-2913
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2913
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2913). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This protocol was approved by the IACUC and IBC Committee in Zhongshan Hospital, Xiamen University (approved number: 2017-015) with a tumor volume less than 5,000 mm3, and conducted in accordance with the Guide for the Care and Use of Laboratory Animal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Senaratna CV, Perret JL, Lodge CJ, et al. Prevalence of obstructive sleep apnea in the general population: A systematic review. Sleep Med Rev 2017;34:70-81. [Crossref] [PubMed]
- Jordan AS, McSharry DG, Malhotra A. Adult obstructive sleep apnoea. Lancet 2014;383:736-47. [Crossref] [PubMed]
- Marshall NS, Wong KK, Cullen SR, et al. Sleep apnea and 20-year follow-up for all-cause mortality, stroke, and cancer incidence and mortality in the busselton health study cohort. J Clin Sleep Med 2014;10:355-62. [Crossref] [PubMed]
- Chung WS, Lin CL. Sleep disorders associated with risk of prostate cancer: a population-based cohort study. BMC Cancer 2019;19:146. [Crossref] [PubMed]
- Seijo LM, Perez-Warnisher MT, Giraldo-Cadavid LF, et al. Obstructive sleep apnea and nocturnal hypoxemia are associated with an increased risk of lung cancer. Sleep Med 2019;63:41-5. [Crossref] [PubMed]
- Almendros I, Montserrat JM, Torres M, et al. Obesity and intermittent hypoxia increase tumor growth in a mouse model of sleep apnea. Sleep Med 2012;13:1254-60. [Crossref] [PubMed]
- Vilaseca A, Campillo N, Torres M, et al. Intermittent hypoxia increases kidney tumor vascularization in a murine model of sleep apnea. PLoS One 2017;12:e0179444. [Crossref] [PubMed]
- Huang MH, Zhang XB, Wang HL, et al. Intermittent hypoxia enhances the tumor programmed death ligand 1 expression in a mouse model of sleep apnea. Ann Transl Med 2019;7:97. [Crossref] [PubMed]
- Almendros I, Montserrat JM, Torres M, et al. Intermittent hypoxia increases melanoma metastasis to the lung in a mouse model of sleep apnea. Respir Physiol Neurobiol 2013;186:303-7. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [Crossref] [PubMed]
- Farh KK, Grimson A, Jan C, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 2005;310:1817-21. [Crossref] [PubMed]
- Barbato S, Solaini G, Fabbri M. MicroRNAs in Oncogenesis and Tumor Suppression. Int Rev Cell Mol Biol 2017;333:229-68. [Crossref] [PubMed]
- Corsini LR, Bronte G, Terrasi M, et al. The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin Ther Targets 2012;16:S103-9. [Crossref] [PubMed]
- Rupaimoole R, Wu SY, Pradeep S, et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun 2014;5:5202. [Crossref] [PubMed]
- Riemann A, Reime S, Thews O. Hypoxia-Related Tumor Acidosis Affects MicroRNA Expression Pattern in Prostate and Breast Tumor Cells. Adv Exp Med Biol 2017;977:119-24. [Crossref] [PubMed]
- Ullmann P, Nurmik M, Begaj R, et al. Hypoxia- and MicroRNA-Induced Metabolic Reprogramming of Tumor-Initiating Cells. Cells 2019;8. [PubMed]
- Santamaria-Martos F, Benitez I, Ortega F, et al. Circulating microRNA profile as a potential biomarker for obstructive sleep apnea diagnosis. Sci Rep 2019;9:13456. [Crossref] [PubMed]
- Gao H, Han Z, Huang S, et al. Intermittent hypoxia caused cognitive dysfunction relate to miRNAs dysregulation in hippocampus. Behav Brain Res 2017;335:80-7. [Crossref] [PubMed]
- Almendros I, Khalyfa A, Trzepizur W, et al. Tumor Cell Malignant Properties Are Enhanced by Circulating Exosomes in Sleep Apnea. Chest 2016;150:1030-41. [Crossref] [PubMed]
- Freitas LS, Silveira AC, Martins FC, et al. Severe obstructive sleep apnea is associated with circulating microRNAs related to heart failure, myocardial ischemia, and cancer proliferation. Sleep Breath 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Almendros I, Wang Y, Becker L, et al. Intermittent hypoxia-induced changes in tumor-associated macrophages and tumor malignancy in a mouse model of sleep apnea. Am J Respir Crit Care Med 2014;189:593-601. [Crossref] [PubMed]
- Ali M, Kowkuntla S, Delloro DJ, et al. Chronic intermittent hypoxia enhances disease progression in myeloma-resistant mice. Am J Physiol Regul Integr Comp Physiol 2019;316:R678-86. [Crossref] [PubMed]
- Zhang XB, Yang YY, Zeng Y, et al. Anti-tumor effect of endostatin in a sleep-apnea mouse model with tumor. Clin Transl Oncol 2019;21:572-81. [Crossref] [PubMed]
- Zhang XB, Zeng YM, Chen XY, et al. Decreased expression of hepatic cytochrome P450 1A2 (CYP1A2) in a chronic intermittent hypoxia mouse model. J Thorac Dis 2018;10:825-34. [Crossref] [PubMed]
- Akbarpour M, Khalyfa A, Qiao Z, et al. Altered CD8+ T-Cell Lymphocyte Function and TC1 Cell Stemness Contribute to Enhanced Malignant Tumor Properties in Murine Models of Sleep Apnea. Sleep 2017;40. [PubMed]
- Almendros I, Montserrat JM, Ramirez J, et al. Intermittent hypoxia enhances cancer progression in a mouse model of sleep apnoea. Eur Respir J 2012;39:215-7. [Crossref] [PubMed]
- Liu R, Zheng H, Li W, et al. Anti-tumor enhancement of Fei-Liu-Ping ointment in combination with celecoxib via cyclooxygenase-2-mediated lung metastatic inflammatory microenvironment in Lewis lung carcinoma xenograft mouse model. J Transl Med 2015;13:366. [Crossref] [PubMed]
- Fehlmann T, Reinheimer S, Geng C, et al. cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin Epigenetics 2016;8:123. [Crossref] [PubMed]
- Mak SST, Gopalakrishnan S, Caroe C, et al. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience 2017;6:1-13. [Crossref] [PubMed]
- Bradley BS, Loftus JC, Mielke CJ, et al. Differential expression of microRNAs as predictors of glioblastoma phenotypes. BMC Bioinformatics 2014;15:21. [Crossref] [PubMed]
- Lai XL, Huang YH, Li YS, et al. Differential expression profiling of microRNAs in para-carcinoma, carcinoma and relapse human pancreatic cancer. Clin Transl Oncol 2015;17:398-408. [Crossref] [PubMed]
- Huang L, Yin ZJ, Feng YF, et al. Identification and differential expression of microRNAs in the ovaries of pigs (Sus scrofa) with high and low litter sizes. Anim Genet 2016;47:543-51. [Crossref] [PubMed]
- Soares CT, Trombone APF, Fachin LRV, et al. Differential Expression of MicroRNAs in Leprosy Skin Lesions. Front Immunol 2017;8:1035. [Crossref] [PubMed]
- Nieto FJ, Peppard PE, Young T, et al. Sleep-disordered Breathing and Cancer Mortality Results from the Wisconsin Sleep Cohort Study. Am J Respir Crit Care Med 2012;186:190-4. [Crossref] [PubMed]
- Chen JC, Hwang JH. Sleep apnea increased incidence of primary central nervous system cancers: a nationwide cohort study. Sleep Medicine 2014;15:749-54. [Crossref] [PubMed]
- Campos-Rodriguez F, Martinez-Garcia MA, Martinez M, et al. Association between Obstructive Sleep Apnea and Cancer Incidence in a Large Multicenter Spanish Cohort. Am J Respir Crit Care Med 2013;187:99-105. [Crossref] [PubMed]
- Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 2009;27:5848-56. [Crossref] [PubMed]
- Nallamshetty S, Chan SY, Loscalzo J. Hypoxia: a master regulator of microRNA biogenesis and activity. Free Radic Biol Med 2013;64:20-30. [Crossref] [PubMed]
- Bandara V, Michael MZ, Gleadle JM. Hypoxia represses microRNA biogenesis proteins in breast cancer cells. BMC Cancer 2014;14:533. [Crossref] [PubMed]
- Belaidi E, Thomas A, Bourdier G, et al. Endoplasmic reticulum stress as a novel inducer of hypoxia inducible factor-1 activity: its role in the susceptibility to myocardial ischemia-reperfusion induced by chronic intermittent hypoxia. Int J Cardiol 2016;210:45-53. [Crossref] [PubMed]
- Almendros I, Wang Y, Gozal D. The polymorphic and contradictory aspects of intermittent hypoxia. Am J Physiol Lung Cell Mol Physiol 2014;307:L129-40. [Crossref] [PubMed]
- Chopra S, Polotsky VY, Jun JC. Sleep Apnea Research in Animals. Past, Present, and Future. Am J Respir Cell Mol Biol 2016;54:299-305. [Crossref] [PubMed]
- Xing B, Li QJ, Li H, et al. miR-140-5p aggravates hypoxia-induced cell injury via regulating MLK3 in H9c2 cells. Biomed Pharmacother 2018;103:1652-7. [Crossref] [PubMed]
- Chen F, Chu L, Li J, et al. Hypoxia induced changes in miRNAs and their target mRNAs in extracellular vesicles of esophageal squamous cancer cells. Thorac Cancer 2020;11:570-80. [Crossref] [PubMed]
- Zhang K, Guo L. MiR-767 promoted cell proliferation in human melanoma by suppressing CYLD expression. Gene 2018;641:272-8. [Crossref] [PubMed]
- Liu F, Hu H, Zhao J, et al. miR-124-3p acts as a potential marker and suppresses tumor growth in gastric cancer. Biomed Rep 2018;9:147-55. [PubMed]
- Yan G, Li Y, Zhan L, et al. Decreased miR-124-3p promoted breast cancer proliferation and metastasis by targeting MGAT5. Am J Cancer Res 2019;9:585-96. [PubMed]
- Hu XX, Feng J, Huang XW, et al. Histone deacetylases up-regulate C/EBPalpha expression through reduction of miR-124-3p and miR-25 in hepatocellular carcinoma. Biochem Biophys Res Commun 2019;514:1009-16. [Crossref] [PubMed]
- Hou ZH, Xu XW, Fu XY, et al. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol 2020;318:C649-63. [Crossref] [PubMed]
- Feng ZY, Xu XH, Cen DZ, et al. miR-590-3p promotes colon cancer cell proliferation via Wnt/beta-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur Rev Med Pharmacol Sci 2017;21:4844-52. [PubMed]
- Sun ZQ, Shi K, Zhou QB, et al. MiR-590-3p promotes proliferation and metastasis of colorectal cancer via Hippo pathway. Oncotarget 2017;8:58061-71. [Crossref] [PubMed]
- Salem M, Shan Y, Bernaudo S, et al. miR-590-3p Targets Cyclin G2 and FOXO3 to Promote Ovarian Cancer Cell Proliferation, Invasion, and Spheroid Formation. Int J Mol Sci 2019;20:1810. [Crossref] [PubMed]
- Salem M, O'Brien JA, Bernaudo S, et al. miR-590-3p Promotes Ovarian Cancer Growth and Metastasis via a Novel FOXA2-Versican Pathway. Cancer Res 2018;78:4175-90. [Crossref] [PubMed]
- Kim CW, Oh ET, Kim JM, et al. Hypoxia-induced microRNA-590-5p promotes colorectal cancer progression by modulating matrix metalloproteinase activity. Cancer Lett 2018;416:31-41. [Crossref] [PubMed]
- Zhang Y, Pan X, Yu X, et al. MicroRNA-590-3p inhibits trophoblast-dependent maternal spiral artery remodeling by repressing low-density lipoprotein receptor-related protein 6. Mol Genet Genomic Med 2018;6:1124-33. [Crossref] [PubMed]
- Bao MH, Li GY, Huang XS, et al. Long Noncoding RNA LINC00657 Acting as a miR-590-3p Sponge to Facilitate Low Concentration Oxidized Low-Density Lipoprotein-Induced Angiogenesis. Mol Pharmacol 2018;93:368-75. [Crossref] [PubMed]
- Olea E, Gaytan SP, Obeso A, et al. Interactions between postnatal sustained hypoxia and intermittent hypoxia in the adulthood to alter brainstem structures and respiratory function. Adv Exp Med Biol 2012;758:225-31. [Crossref] [PubMed]
- Nanduri J, Semenza GL, Prabhakar NR. Epigenetic changes by DNA methylation in chronic and intermittent hypoxia. Am J Physiol Lung Cell Mol Physiol 2017;313:L1096-100. [Crossref] [PubMed]


