
Identification of significant genes in non-small cell lung cancer by bioinformatics analyses
Introduction
Lung cancer is the most commonly diagnosed cancer and leading cause of cancer mortality worldwide, which accounts for 11.6% of total cases and 18.4% of the total deaths (1). Based on histological classification, lung cancer is categorized into non-small cell lung cancer (NSCLC, ~85%) and small-cell lung cancer (SCLC, ~15%), the former group is further classified into three common subtypes, large-cell carcinoma, squamous cell carcinoma, and adenocarcinoma (2,3). Despite progresses achieved in therapies including surgical resection, chemotherapy, radiotherapy, and immunotherapy for NSCLC in recent years, the 5-year survival rate is still low and only 5% (4). Lack of specific molecular biomarker leads to many NSCLC patients were diagnosed at advanced stage, resulting in no long-term survival (5). Encouragingly, with the development of oncogenetics and molecular etiology of lung cancer, great progress has been made in targeted cancer therapy. For example, tyrosine kinase inhibitor (TKI), such as gefitinib and erlotinib, can block the activity of epidermal growth factor receptor (EGFR) reversibly, suppress cell proliferation and transformation, thus improve response rate and prolong survival (6). However, the clinical benefits of these targeted therapies are only restricted to a cohort of NSCLC patients with corresponding targets. Therefore, it is important to further reveal the molecular mechanisms involved in the initiation and progression of NSCLC and to identify the alternated key genes to develop more effective therapies for lung cancer.
Gene chip is a powerful and reliable technologies that can quickly yield quantitative differentially expressed genes (DEGs) and expression profiles by it (7). To date, a large number of microarray data could be explored from the Gene Expression Omnibus public database. With the rapid development of high-throughput sequencing, bioinformatics analysis has been applied in mining the pathophysiological mechanism of different cancers (8-10). In this study, we downloaded 3 NSCLC related mRNA datasets GSE19188, GSE27262, GSE118370 from GEO database to find DEGs, Subsequently, hub genes were found to be associated with survival and further validated when lung tissues compared with adjacent normal tissues. In conclusion, our study can further understand the molecular mechanism of NSCLC and provides potential useful biomarkers for diagnosis, and targeted therapy of NSCLC patients.
Methods
Microarray data
The microarray data GSE19188, GSE27262, GSE118370 used in this study were downloaded from the Gene Expression Omnibus database at NCBI (www.ncbi.nlm.nih.gov/geo/) (11), which is a openly public database. They were all based on the platform of the GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, which consisted of 91 lung cancer and 65 adjacent normal lung tissue, 25 lung cancer and 25 adjacent paired normal lung tissue, 6 lung adenocarcinoma tissues and 6 paired normal lung tissues, respectively. Data processing and identification of DEGs. The DEGs between NSCLC specimen and normal lung specimen were identified via GEO2R, which is an online tool and can be applied to screen DEGs. |logFC| >2 and adjust P<0.05 were considered as cut-off value. Venn software was applied to detect DEGs among the 3 datasets.
Gene ontology (GO) and pathway enrichment analysis of DEGs
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) (12) annotations analysis of DEGs gene were performed via the Database for Annotation, Visualization and Integrated Discovery 6.8 (DAVID6.8) (https://david.ncifcrf.gov/) (13). GO analysis is a commonly useful tool to investigate unique biological properties of DEGs that were involved, including biological processes (BP), cellular components (CC) and molecular function (MF). KEGG is an online database to integrate protein interaction network information and deal with disease, metabolism, biological pathways, and drug research. DAVID, as a comprehensive set of functional annotation tool, can integrate public bioinformatics resources and perform biological analyses of genes by clustering algorithm. P<0.05 was considered significant
Protein-protein interaction (PPI) network and module analysis
Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org) (14) was applied to download the interaction information of human proteins and construct PPI network, then Cytoscape (www.cytoscape.org) (15) was used to visualize PPI network with cut-off criteria of combined score >0.4. In addition, The PPI network modules was analyzed via the Molecular Complex Detection (MCODE) app in Cytoscape based on topology (degree cutoff =2, max. Depth =100, k-core =2, and node score cutoff =0.2).
Survival analysis of crucial genes
Kaplan–Meier plotter (http://kmplot.com/) (16) is a commonly used web tool that is capable to assess the prognostic values of genes in 21 cancer patients, of which the largest dataset including breast, ovarian, lung, and gastric cancer based on GEO, EGA, and TCGA. According to the level of gene expression (high and low), the NSCLC patients were divided into two groups. The HR with 95% confidence intervals and log rank P value were computed and displayed on each plot.
RNA sequencing expression of hub gene in GEPIA
The Gene Expression Profiling Interactive Analysis (GEPIA) is online database that can analyze RNA sequencing expression. To further validate these significantly correlated genes, the GEPIA was used.
Results
Identification of DEGs in lung cancer
We used GEO2R online tool to extract 501, 474, 749 DEGs in GSE19188, GSE27262, GSE118370, of which 357, 329, 359 downregulated and 144, 145, 390 upregulated DEGs, respectively. Then, Veen diagram software was applied to identify the most reliable DEGs among 3 datasets. As shown in Figure 1 and Table 1, in total, 149 DEGs that met the cut-off criteria were obtained, including 127 down-regulated and 22 were up-regulated.
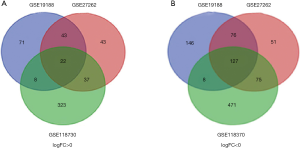
Table 1
| DEGs | Gene names |
|---|---|
| Up-regulated | KIF26B, CCNB1, HMGB3, CD24, CXCL13, GJB2, AURKA, TFAP2A, FERMT1, HMMR, TMPRSS4, HS6ST2, SPP1, SIX1, COL10A1, COL11A1, UGT8, NUF2, MMP1, NEK2, MMP12, CENPF |
| Down-regulated | HBA2///HBA1, RTKN2, EMCN, SOX7, ADARB1, PPP1R14A, WISP2, MFAP4, KCNT2, ERG, SLC6A4, PECAM1, KCNK3, SYNPO2, GIMAP8, OGN, SCARA5, BTNL9, PCAT19, IGSF10, ACVRL1, SCGB1A1, CDO1, CA4, SDPR, TEK, CLIC3, GRK5, DACH1, VGLL3, GUCY1A2, PALM2-AKAP2///AKAP2, STXBP6, S1PR1, EMP2, LYVE1, ADAMTS8, GDF10, LEPROT///LEPR, BCHE, SPOCK2, AKAP12, CD36, PDE5A, LDB2, ROBO4, SPTBN1, CALCRL, CAV1, PPBP, JAM2, PTPRB, QKI, FOXF1, ACADL, ANKRD29, AQP4, PIR-FIGF///FIGF, ITGA8, MT1M, TNNC1, IL1RL1, FAT3, MCEMP1, HBB, FHL1, RHOJ, THBD, KLF4, SCN7A, FMO2, ABCA8, MYZAP, AOC3, SFTPC, ADRB1, SEMA3G, TCF21, TGFBR3, HHIP, ADH1B, ARHGEF26, ARHGAP6, LINC00968, ASPA, CCL15-CCL14///CCL14, FABP4, EDNRB, SCN4B, FCN3, MYCT1, KANK3, STX11, LINC00312, CCDC85A, FAM107A, CCBE1, PGM5, GPX3, AGER, RGCC, VWF, MARCO, SEMA5A, ABI3BP, CD93, TIE1, KIAA1462, VIPR1, AGTR1, EPAS1, RAMP3, CLIC5, SLIT2, FHL5, ADAMTSL3, CLDN18, C2orf40, CDH5, PDK4, GPM6A, COL6A6, ANGPT1, SMAD6, TMEM100, DUOX1, AFF3 |
GO and KEGG pathway analysis of DEGs
To further identify the potential biological functions of these 149 DEGs, DAVID online software was used to analyze GO categories. The results of GO functional enrichment analysis, as shown in Table 2, indicated that, as for BP, upregulated DEGs were significantly enriched in collagen catabolic process, sensory perception of sound, G2M transition of mitotic cell cycle, cell division, inner ear morphogenesis, and downregulated DEGs in angiogenesis, cell adhesion, vasculogenesis, single organismal cell-cell adhesion, response to hypoxia; for cell composition (CC) part, upregulated DEGs were particularly involved in centrosome, proteinaceous extracellular matrix, collagen trimer, spindle pole and downregulated genes in membrane raft, proteinaceous extracellular matrix, cell surface, plasma membrane, integral component of plasma membrane, external side of plasma membrane; in the MF section, the upregulated DEGs participated in extracellular matrix binding, serine-type endopeptidase activity and downregulated genes in heparin binding, receptor activity, transformation growth factor beta binding, peroxidase activity. All terms are closely associated with the tumorigenesis and development. On the other hand, KEGG pathway enrichment analysis was performed to analyze the biological functions of these genes. The most enriched KEGG pathways were as follows: ECM–receptor interaction, Vascular smooth muscle contraction, PPAR signaling pathway, Adrenergic signaling in cardiomyocytes, cell adhesion molecules (CAMs) and focal adhesion (Table 3).
Table 2
| Expression | Category | Term | Count | P value | FDR |
|---|---|---|---|---|---|
| Upregulated | GOTERM_BP_DIRECT | GO:0030574~collagen catabolic process | 4 | 6.69E-05 | 0.088526 |
| GOTERM_BP_DIRECT | GO:0007605~sensory perception of sound | 4 | 5.82E-04 | 0.768065 | |
| GOTERM_BP_DIRECT | GO:0000086~G2/M transition of mitotic cell cycle | 4 | 6.34E-04 | 0.837064 | |
| GOTERM_BP_DIRECT | GO:0051301~cell division | 5 | 8.39E-04 | 1.104883 | |
| GOTERM_BP_DIRECT | GO:0042472~inner ear morphogenesis | 3 | 0.001902 | 2.490103 | |
| GOTERM_CC_DIRECT | GO:0005813~centrosome | 5 | 0.001285 | 1.321892 | |
| GOTERM_CC_DIRECT | GO:0005578~proteinaceous extracellular matrix | 4 | 0.003438 | 3.501323 | |
| GOTERM_CC_DIRECT | GO:0005581~collagen trimer | 3 | 0.004973 | 5.029117 | |
| GOTERM_CC_DIRECT | GO:0000922~spindle pole | 3 | 0.006912 | 6.926107 | |
| GOTERM_CC_DIRECT | GO:0045120~pronucleus | 2 | 0.00804 | 8.014669 | |
| GOTERM_MF_DIRECT | GO:0050840~extracellular matrix binding | 2 | 0.030374 | 27.11998 | |
| GOTERM_MF_DIRECT | GO:0004252~serine-type endopeptidase activity | 3 | 0.036114 | 31.42547 | |
| Down regulated | GOTERM_BP_DIRECT | GO:0001525~angiogenesis | 13 | 2.78E-08 | 4.38E-05 |
| GOTERM_BP_DIRECT | GO:0007155~cell adhesion | 15 | 1.99E-06 | 0.003144 | |
| GOTERM_BP_DIRECT | GO:0001570~vasculogenesis | 5 | 5.10E-04 | 0.800955 | |
| GOTERM_BP_DIRECT | GO:0016337~single organismal cell-cell adhesion | 6 | 5.52E-04 | 0.866402 | |
| GOTERM_BP_DIRECT | GO:0001666~response to hypoxia | 7 | 9.87E-04 | 1.544895 | |
| GOTERM_CC_DIRECT | GO:0045121~membrane raft | 10 | 6.32E-06 | 0.007526 | |
| GOTERM_CC_DIRECT | GO:0005578~proteinaceous extracellular matrix | 11 | 7.68E-06 | 0.009144 | |
| GOTERM_CC_DIRECT | GO:0009986~cell surface | 15 | 8.11E-06 | 0.009649 | |
| GOTERM_CC_DIRECT | GO:0005886~plasma membrane | 46 | 4.84E-05 | 0.057592 | |
| GOTERM_CC_DIRECT | GO:0005887~integral component of plasma membrane | 23 | 6.57E-05 | 0.078202 | |
| GOTERM_CC_DIRECT | GO:0009897~external side of plasma membrane | 8 | 4.07E-04 | 0.483305 | |
| GOTERM_MF_DIRECT | GO:0008201~heparin binding | 7 | 5.16E-04 | 0.682835 | |
| GOTERM_MF_DIRECT | GO:0004872~receptor activity | 7 | 0.002471 | 3.232508 | |
| GOTERM_MF_DIRECT | GO:0050431~transforming growth factor beta binding | 3 | 0.004425 | 5.719841 | |
| GOTERM_MF_DIRECT | GO:0004601~peroxidase activity | 3 | 0.008313 | 10.493 |
Table 3
| Pathway ID | Name | Count | P value | Genes | FDR |
|---|---|---|---|---|---|
| hsa04512 | ECM-receptor interaction | 7 | 1.70E-04 | VWF, CD36, COL6A6, ITGA8, COL11A1, SPP1, HMMR | 0.189451 |
| hsa04270 | Vascular smooth muscle contraction | 5 | 0.025531 | RAMP3, AGTR1, GUCY1A2, CALCRL, PPP1R14A | 25.03342 |
| hsa03320 | PPAR signaling pathway | 4 | 0.026133 | CD36, FABP4, ACADL, MMP1 | 25.54754 |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 5 | 0.042961 | AGTR1, ADRB1, TNNC1, SCN4B, SCN7A | 38.68835 |
| hsa04514 | Cell adhesion molecules (CAMs) | 5 | 0.046888 | CLDN18, ITGA8, PECAM1, JAM2, CDH5 | 41.43357 |
| hsa04510 | Focal adhesion | 6 | 0.047002 | VWF, CAV1, COL6A6, ITGA8, COL11A1, SPP1 | 41.51176 |
Construction PPI network and modular analysis
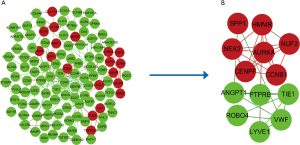
To further predict the interaction of the DEGs at the protein level, the PPI network was constructed, in which 102 DEGs were imported into and 47 were not contained totally. The constructed PPI network contained 204 interaction pairs. Subsequently, cytotype MCODE app was employed to identify modules. As displayed in Figure 2, the top 1 significant module included 13 central nodes among the 102 nodes. Among 13 central nodes, 7 including CCNB1, AURKA, HMMR, SPP1, NUF2, NEK2, CENPF were upregulated and 6 including LYVE1, ROBO4, PTPRB, VWF, TIE1, ANGPT1 were downregulated.
Analysis of core genes by the Kaplan Meier plotter and GEPIA
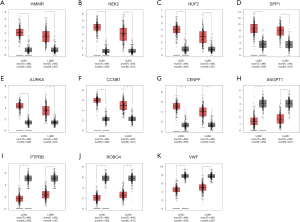
In an attempt to gain insight into association between hub genes and NSCLC patients, Kaplan Meier plotter was utilized to predict the prognostic value of 13 core genes survival data. The result revealed that 11 DEGs had a significant survival while 2 had no significance (P<0.05, Table 4 & Figure 3). ROBO4 with low expression was associated with better overall survival for NSCLC patients, as well as PTPRB, VWF, ANGPT1 (P<0.05). Additionally, high expression of CCNB1 was associated with poorer overall survival, as well as CCNB1, AURKA, HMMR, SPP1, NUF2, NEK2, CENPF (P<0.05). Moreover, to further verify the expression of these DEGs, GEPIA was employed to dig up the 11 gene expression level between lung cancer and normal people. As graphed in Table 5 & Figure 4, notably, the results were in line with the survival analysis above, which imply that the expression levels of the 11 hub genes are particularly associated with clinical prognosis of NSCLC patients and they may play vital roles in the progression of NSCLC.
Table 4
| Category | Genes |
|---|---|
| Genes with significantly better survival (P<0.05) | ROBO4, PTPRB, VWF, ANGPT1 |
| Genes with significantly worse survival (P<0.05) | CCNB1, AURKA, HMMR, SPP1, NUF2, NEK2, CENPF |
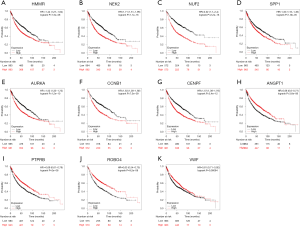
Table 5
| Category | Genes |
|---|---|
| Genes with high expressed in LC (P<0.05) | ROBO4, PTPRB, VWF, ANGPT1 |
| Genes with low expressed in LC (P<0.05) | CCNB1, AURKA, HMMR, SPP1, NUF2, NEK2, CENPF |
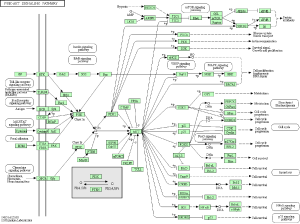
KEGG pathway enrichment of 11 genes reanalysis
KEGG pathway was re-analyzed to investigate the possible pathway of 11 genes. Enrichment analysis showed that the module genes were mainly associated with ECM-receptor interaction and PI3K-Akt signaling pathway (Table 6 & Figure 5).
Table 6
| Pathway ID | Name | Count | P value | Genes | FDR |
|---|---|---|---|---|---|
| cfa04512 | ECM-receptor interaction | 3 | 0.00238 | VWF, SPP1, HMMR | 1.67928 |
| cfa04151 | PI3K-Akt signaling pathway | 3 | 0.03261 | VWF, ANGPT1, SPP1 | 20.9804 |
Discussion
At present, the diagnosis and treatment of NSCLC is still far from satisfactory, and the number of this case is still rising year by year. It is necessary to investigate the pathogenesis and biomarker of NSCLC to provide effective treatment. Great progress has been made on the mechanism of initiation and development of NSCLC. Many experiments including vitro tumor cell lines, animal tumor models, and patients’ tumor model have been done, however, NSCLC demands more comprehensive analysis because the progress of lung cancer is a multi-stage and multi-cause process. Fortunately, with the development of human genome sequencing, the high throughput and associated tumor database developed and were more available to get. The integration of data by bioinformatics analyses from multiple datasets has become a vital source of data for studies of lung cancer. For example, using GSE19804 dataset, Yang et al. identified hub genes including UBE2C, DLGAP5, TPX2, CCNB2, BIRC5, KIF20A, TOP2A, GNG11, and ANXA1 associated with prognosis in nonsmoking females with NSCLC patients (17). Similarly, using 4 dataset GSE21933, GSE33532, GSE44077 and GSE74706, CCNB1, CCNA2, CEP55, PBK and HMMR was identified and associated with poorer survival (18).
In the current study, we attempted to identify tumor related genes that contribute to NSCLC overall survival via series of database. We used bioinformatical methods based on 3 profile datasets (GSE19188, GSE27262 and GSE118370). One hundred and twenty-two lung cancer specimens and 96 normal specimens were enrolled in this research. In particular, we were able to validate 11 genes that significantly associated with prognosis. First, we extracted 149 common DEGs yielded from 3 datasets (|logFC| >2 and adjust P value <0.05), the vast majority were down-regulated, of which including 127 downregulated and 22 upregulated genes. Next, we performed GO and KEGG pathway functional enrichment by DAVID online tool on these DEGs. By performing with GO enrichment analysis, the DEGs were mainly involved in angiogenesis, cell adhesion, vasculogenesis and collagen catabolic process, all these important biological progresses processes participated in the pathophysiological mechanism of NSCLC. Angiogenesis, one of hallmarks of cancer acquired during the multistep development of human tumor (19). A study showed that angiogenic switch is always activated and remains on, resulting in new vessels sprout from quiescent vasculature to help sustain neoplastic growths during tumor progression (20). As for GO cell component (CC), the DEGs were enrich in centrosome, proteinaceous extracellular matrix, plasma membrane, integral component of plasma membrane, cell surface, proteinaceous extracellular matrix and for MF, the DEGs were significantly involved in the heparin binding, extracellular matrix binding, serine-type endopeptidase activity. KEGG pathway enrichment analysis revealed that DEGs are mainly concentrated in the ECM-receptor interaction, Vascular smooth muscle contraction, PPAR signaling pathway. The pathways of ECM-receptor interaction is important mediators of growth, proliferation, survival, angiogenesis and migration of cancer (21), consistent with the results obtained in this study. In addition, we constructed PPI modules and identified 13 high interrelated nodes by mocode app. Subsequently, we performed survival of 13 genes and identified 11 related gene that significantly correlated prognosis analysis in NSCLC patients. Of the 11 genes identified, 7 genes with high expression indicated worse survival, but other 6 genes with low expression indicated better survival. In validating these 11 genes, GEPIA was applied and all genes make sense when lung cancer samples compared with normal samples. Finally, we re-analyzed 11 genes via DAVID for KEGG enrichment and found that 3 genes (VWF, SPP1, and HMMR) enriched in ECM-receptor interaction and 3 genes (VWF, ANGPT1, and SPP1) enriched in PI3K-Akt signaling pathway had a significance (P<0.05). We are particularly interested in VWF and SPP1, because they are common genes in two pathways.
VWF, Von Willebrand factor, a large multimeric plasma glycoprotein originated from endothelial cells, platelets and megakaryocytes. It has been widely known as its function in haemostasis to enables capture of platelets at sites of endothelial damage (22,23), and the function of promoting angiogenesis (24). Recent advances revealed that GATA3 can induce VWF upregulation in the lung adenocarcinoma vasculature by binding to the +220 GATA binding motif on the human VWF promoter (25) and plasma VWF/ADAMTS-13 ratio may act as an independent predictive factor for mortality in patients with advanced NSCLC (26). Another study indicated that VWF with low expression in osteosarcoma tumors can potentially contribute to metastasis (27).
SPP1, secreted phosphoprotein 1, also called OPN. It is located on chromosome 4 in locus 4q13.22 and encoded by the human gene SPP1 (28) that include seven exons and can be alternatively spliced to produce different variants (29). It can be produced by osteoclasts, endothelial cells, epithelial cells, and immune cells to play a vital role in normal and disease BP, including bone remodeling, immune regulation (30) and cell adhesion (31). It can bind to integrins and CD44, resulting in inflammatory disorders, autoimmune diseases, and tumorigenesis (30). In non-small cell lung cancers (NSCLC), SPP1 induces VEGF expression and promotes tumor progression (12). Altogether, it can be a useful target and potential therapy target. Numerous studies have demonstrated that these two genes were related to distinct types of cancer, however, few papers have been studied in lung cancer. Also, CENPF, PTPRB, and NUF2 are rarely reported after we searched these genes in PubMed online website. Taken together, our study linked to NSCLC pathogenesis could improve the understanding of underlying molecular mechanisms of NSCLC and provide useful information for future study of new anticancer in lung cancer.
Conclusions
We identified DEGs between lung cancer and normal tissues on the via bioinformatics analysis and the results revealed they may play crucial roles in the progression of lung cancer; however, Further experiments are needed to verify these predictions. Anyway, this study may provide some potential biomarkers and targets for NSCLC diagnosis and therapy.
Acknowledgments
Funding:
Availability of data and materials: The datasets generated and/or analyzed during the study are available from the corresponding author upon reasonable request.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2596). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Torre LA, Siegel RL, Jemal A. Lung Cancer Statistics. Adv Exp Med Biol 2016;893:1-19. [Crossref] [PubMed]
- Ridge CA, McErlean A, Ginsberg M. Epidemiology of Lung Cancer. Semin Intervent Radiol 2013;30:93-8. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Keith RL, Miller YE. Lung cancer chemoprevention: current status and future prospects. Nat Rev Clin Oncol 2013;10:334-43. [Crossref] [PubMed]
- Lee VH, Tin VP, Choy TS, et al. Association of exon 19 and 21 EGFR mutation patterns with treatment outcome after first-line tyrosine kinase inhibitor in metastatic non-small-cell lung cancer. J Thorac Oncol 2013;8:1148-55. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer Genome Landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Pan JH, Zhou H, Cooper L, et al. LAYN Is a Prognostic Biomarker and Correlated With Immune Infiltrates in Gastric and Colon Cancers. Front Immunol 2019;10:6. [Crossref] [PubMed]
- Jia D, Li S, Li D, et al. Mining TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY) 2018;10:592-605. [Crossref] [PubMed]
- Falzone L, Lupo G, La Rosa GRM, et al. Identification of Novel MicroRNAs and Their Diagnostic and Prognostic Significance in Oral Cancer. Cancers (Basel) 2019;11:610. [Crossref] [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009;37:D885. [Crossref] [PubMed]
- Ogata H, Goto S, Sato K, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;27:29-34.
- Huang da W. Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Kohl M, Wiese S, Warscheid B. Cytoscape: Software for Visualization and Analysis of Biological Networks. Methods Mol Biol 2011;696:291-303. [Crossref] [PubMed]
- Győrffy B, Surowiak P, Budczies J, et al. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PloS one 2013;8:e82241. [Crossref] [PubMed]
- Yang G, Chen Q, Xiao J, et al. Identification of genes and analysis of prognostic values in nonsmoking females with non-small cell lung carcinoma by bioinformatics analyses. Cancer Manag Res 2018;10:4287-95. [Crossref] [PubMed]
- Xiao Y, Feng M, Ran H, et al. Identification of key differentially expressed genes associated with nonsmall cell lung cancer by bioinformatics analyses. Mol Med Rep 2018;17:6379-86. [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Cubillo A, Álvarez-Gallego R, Muñoz M, et al. Dynamic Angiogenic Switch as Predictor of Response to Chemotherapy-Bevacizumab in Patients With Metastatic Colorectal Cancer. Am J Clin Oncol 2019;42:56-9. [Crossref] [PubMed]
- Zhang HJ, Tao J, Sheng L, et al. RETRACTED: Twist2 promotes kidney cancer cell proliferation and invasion via regulating ITGA6 and CD44 expression in the ECM-Receptor-Interaction pathway. Biomed Pharmacother 2016;81:453-9. [Crossref] [PubMed]
- Lenting PJ, Pegon JN, Groot E, et al. Regulation of von Willebrand factor-platelet interactions. Thromb Haemost 2010;104:449-55. [Crossref] [PubMed]
- Hassan MI, Saxena A, Ahmad F. Structure and function of von Willebrand factor. Blood Coagul Fibrinolysis 2012;23:11-22. [Crossref] [PubMed]
- Lenting PJ, Casari C, Christophe OD, et al. von Willebrand factor: the old, the new and the unknown. J Thromb Haemost 2012;10:2428-37. [Crossref] [PubMed]
- Xu Y, Pan S, Liu J, et al. GATA3-induced vWF upregulation in the lung adenocarcinoma vasculature. Oncotarget 2017;8:110517-29. [Crossref] [PubMed]
- Guo R, Yang J, Liu X, et al. Increased von Willebrand factor over decreased ADAMTS-13 activity is associated with poor prognosis in patients with advanced non-small-cell lung cancer. J Clin Lab Anal 2018;32:e22219. [Crossref] [PubMed]
- Eppert K, Wunder JS, Aneliunas V, et al. von Willebrand factor expression in osteosarcoma metastasis. Mod Pathol 2005;18:388-97. [Crossref] [PubMed]
- Sarosiek K, Jones E, Chipitsyna G, et al. Osteopontin (OPN) Isoforms, Diabetes, Obesity, and Cancer; What Is One Got to Do with the Other? A New Role for OPN. J Gastrointest Surg 2015;19:639-50. [Crossref] [PubMed]
- Yamamoto S, Hijiya N, Setoguchi M, et al. Structure of the osteopontin gene and its promoter. Ann N Y Acad Sci 1995;760:44-58. [Crossref] [PubMed]
- Hao C, Cui Y, Owen S, et al. Human osteopontin: Potential clinical applications in cancer Int J Mol Med 2017;39:1327-37. (Review). [Crossref] [PubMed]
- Maeda N, Maenaka K. The Roles of Matricellular Proteins in Oncogenic Virus-Induced Cancers and Their Potential Utilities as Therapeutic Targets. Int J Mol Sci 2017;18:2198. [Crossref] [PubMed]


