
MicroRNA-7 inhibits hepatocellular carcinoma cell invasion and metastasis by regulating Atg5-mediated autophagy
Introduction
Hepatocellular carcinoma (HCC) is characterized by high morbidity, poor prognosis, and strong invasiveness. It is one of the most common malignant tumors of the digestive system. To date, there have been many important advances in the research of liver cancer in medicine. However, the 5-year survival rate of patients with liver cancer is low (5% to 9%) (1). MicroRNA (miR) is a small non-coding single-stranded RNA composed of 19–25 nucleotides. The abnormal expression of miRNA in liver cancer cells may play an important role in the pathophysiology of liver cancer. Autophagy is a catabolic process (2-6). Autophagy vesicles can isolate damaged macromolecules from organelles, and are degraded and recycled after lysosome fusion (7-10). Autophagy-related genes and proteins (Atg) are at the core of autophagy regulation (11). microRNA-7 (miR-7) is considered to be a tumor suppressor miRNA in a lot of cancers such as breast, brain, head and neck, liver, colon and melanoma. The ability of tumor invasion and metastasis is an important indicator for evaluating the degree of malignancy of HCC, and autophagy may affect the invasion and metastasis ability of tumor cells (12-17). The purpose of this study was to investigate the effect of miR-7 on targeting autophagy-related protein Atg5 in inhibiting the ability of autophagy cause invasion and metastasis of liver cancer cells.
Methods
Material
Human liver cancer specimens (all HCC) and normal liver tissues adjacent to the cancer 25 patients who underwent surgery from June 2019 to December 2019 (31–67 years old, 16 males and 9 females) were reviewed in Department of Pathology, Third Xiangya Hospital, Central South University. The SMMC-7721 human liver cancer cell line was purchased from Shanghai Cell Bank, Chinese Academy of Sciences (Xuhui District, Shanghai, China). Serum was purchased from Gibco Corporation (Grand Island, NY, USA). A quantitative real-time polymerase chain (qRT-PCR) kit was purchased from Beijing Quanshijin Biotechnology Co, Ltd. (Dongsheng International Science Park, Zhongguancun, Haidian District, Beijing). Rabbit anti-mouse LC3, Atg5, N-cadherin (calcium adhesion molecule N), vimentin, snail (snail protein), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies, and goat anti-rabbit horseradish peroxidaselabeled secondary antibodies were purchased from CST China Company (Shanghai, China). GFP-RFP-LC3 autophagy double-labeled adenovirus was purchased from Shanghai Hanheng Biotechnology Company (Pudong New District, Shanghai, China). miR-7 mimic and inhibitor were purchased from Guangzhou Ruibo Biological Technology Co., Ltd (Guangzhou, China). 3-MA was purchased from Sigma-Aldrich Corp (St. Louis, MO, USA). Transwell Room and Matrigel glue were purchased from BD Corporation Shanghai Co, Ltd (Shanghai, China). The trial was conducted in accordance with the Declaration of Helsinki. All patients signed an informed consent form, and this study was approved by the Ethics Committee of the Third Xiangya Hospital of Central South University (No. 2020-S053).
Methodology
qRT-PCR to detect the expression level of miR-7 and Atg5 in human normal liver tissues and liver cancer tissues
Total RNA was extracted from liver cancer tissues and normal liver tissues using the Trizol method and reverse-transcribed into cDNA through specific preprimers and postprimers. The tissues were then predenatured at 94 °C for 10 min and 94 °C for 1 min, and then annealed at 60 °C for 1 min, extended at 72 °C for 5 min, amplified for 30 cycles, and finally extended at 72 °C for 10 min . Quantitative analysis of the results in each group was performed using the comparison threshold method with the following formula: quantitative copy number of target gene = 2−△△CT; △CT control group = CT target gene-CT internal reference; △△CT = △CT experimental group − △CT control group.
Cell transfection, model of cell ischemia, and 3-MA treatment
For routine cultivation, cells were recovered and placed in an incubator at 37 °C with 5% CO2. Transfection was carried out according to the operation manual provided by the manufacturer. The miR-7 mimic and inhibitor were dissolved to the appropriate concentration according to the instructions. The transfection reagent Lipo3000 was used for integration with the miR-7 mimic, and inhibitor was added to the 6-well plate to continue routine cultivation. The transfected cells were further cultured in an incubator at 37 °C with 5% CO2. After 4–6 h, the solution was replaced with 10% fresh medium, and cultivation was continued for 48 h. The culture medium was discarded, 2 mL of Hank’s culture medium was added to each well, and culturing was continued in order to achieve cell starvation and ischemia. After 1 h, Hank’s culture solution was discarded, 2 mL of 10% fresh medium was added, and cells were placed in an incubator at 37 °C with 5% CO2 to continue culturing for 12 h.
Test of cell proliferation ability
When cultured to cover 80–90% of the area of each well, the cells were digested and resuspended by trypsin. After counting, the cell concentration was adjusted to 1,000 cells/mL. The cell suspension was added to a 6-well plate filled with complete medium. After culturing for 2 weeks, and when there were obvious cell colonies visible to the naked eye, the culture was terminated and the medium was discarded. This was followed by washing 3 times with phosphate-buffered saline (PBS), after which the number of clones per well was counted after staining with 0.5% crystal violet.
Detection of the invasion and metastasis of liver cancer cells
The cultured cells were digested and resuspended when they covered 80–90% of the area of the well. These cells were then counted, and the concentration was adjusted to 105 cells/mL. Whole medium (600 µL/well) was added to the 24-well plate. Transwell chambers, both with a Matrigel gel chamber to observe the invasion ability of SMMC-7721 cells and without a Matrigel gel chamber to observe the tumor cell transfer ability, were put into a 24-well plate, and 100 µL of cell suspension of each group was added to the chamber. These sets were incubated for 24–48 h. Next, the medium was discarded, and cells were taken out the chamber, stained with 0.1% crystal violet staining solution at room temperature for 3 h, observed, and counted under the microscope.
LC3 autophagy double-labeled adenovirus (GFP-RFP-LC3) transfection to detect the formation of autophagosomes
The cells were cultured to about 70% of the area of well, and then infected by adenovirus. After 4–6 h, the cell medium was discarded and cultured with fresh medium again in the incubator for 24–48 h. After simulated cell ischemia and hypoxia treatment, the autophagosomes were observed under an inverted fluorescence microscope.
Western blot to detect the expression of the target protein
Lysate was added into each group, and the supernatant was taken after centrifugation at 12,000 r/min and a centrifugal radius of 6 cm for 15 min at 4 °C. Protein was quantified by bicinchoninic acid (BCA) method, with the protein being denatured in a 100 °C water bath. Next, 50 µg/well of protein in each group was taken for gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membrane. Then, 5% non-fat milk powder was sealed at room temperature for 1 h, and primary antibody was applied (rabbit anti-mouse LC3, Atg5, N-cadherin, vimentin, snail, and GAPDH, 1:1,000), shaken at 4 °C overnight, and washed 3 times with tris-buffered saline with Tween20 (TBST). Sheep anti-rabbit horseradish peroxidase–labeled secondary antibody was applied (1:5,000) for 1 h at room temperature, and luminescent solution was added to the PVDF membrane. The picture was exposed and saved, and a gel image processing system was used to analyze the molecular weight and net optical density value of the target band.
Statistical analysis
The experimental data are expressed as (mean ± standard deviation). SPSS22.0 statistical software was used to conduct the Student’s t-test and one-way analysis of variance. A P value <0.05 was considered statistically significant.
Results
Downregulation of miR-7 expression in human HCC and its correlation with Atg5
By using qRT-PCR to detect the expression of miR-7 and Atg5 in normal human liver tissues and liver cancer tissues, we found that miR-7 expression in liver cancer tissue (16.72±4.71) was significantly lower than that in normal liver tissue (24.85±4.83, t1=6.02; P<0.05) (see Figure 1A), while the expression of Atg5 in liver cancer tissue (13.70±2.80) was higher than that in normal liver tissue (8.68±2.79, t2=6.36; P<0.05) (see Figure 1B). Furthermore, miR-7 and Atg5 were highly correlated in liver cancer tissues and were negatively correlated (r=−0.97) (Figure 1C). This indicates that miR-7 may have an inhibitory effect on liver cancer, which may be exerted by negatively regulating Atg5.

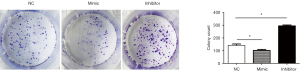
MiR-7 inhibits the proliferation of liver cancer cells
SMMC-7721 liver cancer cells were divided into 3 groups: the NC group (for the blank control of transfection of liver cancer cells), the mimic group (for miR-7 specific agonist mimic transfection of liver cancer cells), and the inhibitor group (for liver cancer cells transfected with a specific inhibitor of miR-7, and then tested to determine the effect of miR-7 on the proliferative ability of 7,721 hepatoma cells by plate cloning experiment). The experimental results showed that the miR-7 expression level in the mimic group increased, and the number of hepatoma cell proliferative cells decreased compared with the NC group (t1=3.22, *P<0.05). The miR-7 expression level decreased, and the number of proliferative liver cancer cells was higher than that of the NC group (t2=12.47, P<0.05) and mimic group (t3=17.64, P<0.05) (Figure 2). The experimental results suggest that miR-7 may have the effect of inhibiting the proliferation of liver cancer cells.
Overexpression of miR-7 can downregulate Atg5 to inhibit autophagy, and low expression of miR-7 can upregulate Atg5 to enhance autophagy
After transfection with LC3 autophagy double-labeled adenovirus, the formation of autophagosomes in liver cancer cells was observed under a fluorescent microscope, and the expression of autophagy-related protein LC3II/I was detected by immunoblot to determine the effect of change of miR-7 expression on intracellular autophagy in liver cancer. The results showed that the number of autophagosomes in the mimic group was significantly reduced compared with the NC group (5.5±1.05 vs. 12.83±1.47, t=9.94, P<0.05) and the inhibitor group (36.33±2.16, F=588.68, P<0.05, Figure 3A). Western blot results showed that the expression levels of Atg5 and LC3II (active LC3) in the mimic group were lower than those in the NC group, while the expression levels of Atg5 and LC3II in the inhibitor group were significantly higher than those in the mimic and NC groups (see Figure 3B, Table 1). These results indicate that miR-7 overexpression in hepatoma cells could reduce Atg5 expression, thereby inhibiting HCC autophagy. Conversely, miR-7 overexpression enhanced Atg5 expression and autophagy, indicating that miR-7 can inhibit autophagy by downregulating Atg5 in hepatoma cells.
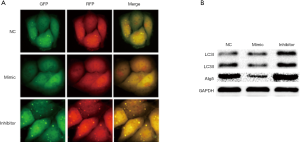
Table 1
| Groups | LC3 I | LC3 II | ATG5 |
|---|---|---|---|
| NC | 0.38±0.06 | 0.74±0.11 | 0.97±0.04 |
| Mimic | 0.68±0.13 | 0.51±0.06 | 0.49±0.07 |
| Inhibitor | 1.08±0.11 | 0.93±0.12 | 1.93±0.11 |
| F value | 55.76* | 23.58* | 395.26* |
*, P<0.05.
Inhibition of autophagy can increase the inhibitory effect of miR-7 on the invasion and metastasis of liver cancer cells
Hepatocarcinoma cells were divided into 3 groups: NC group, the mimic group, and the mimic + 3-MA group (transfected mimic plus autophagy inhibitor 3-MA with a final concentration of 10 mmol/L). The invasion and metastasis ability of liver cancer was detected by Transwell experiment, and the expression level of epithelial-mesenchymal transition (EMT)-related proteins were detected by Western blot. Transwell results showed that the mimic group (352.50±28.65) was less invasive and metastatic than the NC group (576.17±63.04); the mimic + 3-MA group (134.33±24.87) was also less invasive and metastatic than the mimic group (P<0.05) (Figure 4A). Western blot results showed that the expression of EMT-related protein in the mimic group was lower than that in the NC group, while the expression of EMT-related protein in the mimic + 3-MA group was lower than that in the mimic group and significantly lower than that in the NC group (Figure 4B, Table 2). The results show that inhibition of autophagy could increase the inhibitory effect of miR-7 on the invasion and metastasis ability of liver cancer cells, and enhance the anti-tumor effect of miR-7.
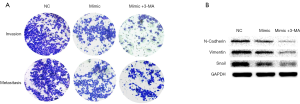
Table 2
| Groups | N-CA | Vimentin | Snail |
|---|---|---|---|
| NC | 0.73±0.12 | 0.86±0.07 | 1.23±0.21 |
| Mimic | 0.37±0.04 | 0.60±0.07 | 0.54±0.07 |
| Inhibitor | 0.16±0.05 | 0.33±0.08 | 0.31±0.05 |
| F value | 66.55* | 72.75* | 67.16* |
*, P<0.05. EMT, epithelial-mesenchymal transition.
Discussion
Previous studies have shown that microRNA as a tumor suppressor or oncogene is involved in the pathophysiological process of liver cancer, indicating that microRNA may be a potential marker for the clinical diagnosis, treatment, and prognosis of liver cancer (18-24). Fang et al. reported that miR-7 could regulate PIK3CD expression through PI3K/AKT/mTOR pathway, thereby inhibiting the survival of liver cancer cells (23). In this study, we found that miR-7 has a low expression in liver cancer tissues, and the overexpression of miR-7 could significantly increase the inhibitory effect on tumor proliferation, invasion, and metastasis, suggesting that miR-7 may have anticancer effects. At the same time, it was found that Atg5 was highly expressed in liver cancer tissues, indicating that Atg5 may be related to tumorigenesis and progression. We also found that miR-7 and Atg5 were highly negatively correlated in liver cancer tissues, suggesting that miR-7 may play an antitumor role by downregulating Atg5 in liver cancer tissues.
The miRNA in liver cancer cells is closely related to autophagy, and miRNA can regulate autophagy through various mechanisms. Ge et al. found that miR-100 promoted Atg7-dependent autophagy by directly inhibiting the mTOR and IGF-1R signaling pathways in liver cancer cells, which in turn affected liver cancer progression (25). Stiuso et al. investigated the effect of sorafenib on the treatment of liver cancer and found that the expression of miR-423-5p was upregulated after sorafenib therapy, which in turn activated autophagy, inhibited the proliferation of hepatoma cells and the cell cycle (26). Zhou et al. found that miR-185 could activate autophagy by targeting AKT1, RHEB, and RICTOR in liver cancer cells, and finally induced an antitumor effect (27). In this study, we found that miR-7 could negatively regulate Atg5 and downregulate autophagy in hepatoma cells while the overexpression of miR-7 could lower the level of autophagy in hepatoma cells. Meanwhile, a low expression of miR-7 increasing the high level of autophagy suggested that miR-7can regulate the autophagy of liver cancer cells through Atg5.
Although the autophagy mechanism in the occurrence and development of liver cancer has been widely studied, the true role of autophagy remains unclear (28). It is currently believed that autophagy has a dual role in the progression of liver cancer: that is, it is involved in tumorigenesis and tumor suppression. Chang et al. have found that miR-375 inhibits autophagy by targeting Atg7, and ultimately reduces the activity of liver cancer cells under hypoxia (29). It has also been found that autophagy under hypoxic conditions can protect the energy supply of liver cancer cells during proliferation and invasion, thereby promoting the survival of liver cancer cells. Wang et al. used Torin-2 in 3 cell lines, Hep G2, SNU-182, and Hep 3B2.1-7 to specifically block mTORC1 activity, which enhanced autophagy and ultimately inhibited liver cancer cell proliferation (30). Through experimental research, we found that inhibition of autophagy in liver cancer cells could inhibit the invasion and metastasis of liver cancer and reduce the expression of EMT-related proteins, indicating that autophagy has an antitumor effect.
In general, miR-7 shows low expression in hepatoma cells and can downregulate Atg5 to inhibit autophagy, ultimately inhibiting the proliferation, invasion, and metastasis of hepatoma cells. Therefore, miR-7 is expected to be an effective indicator for clinical treatment of liver cancer and a predictor of prognosis. The regulation of autophagy by miR-7 represents a novel approach in the treatment of liver cancer and the prediction of prognosis.
Acknowledgments
Funding: None.
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1930
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1930). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients signed an informed consent form, and this study was approved by the Ethics Committee of the Third Xiangya Hospital of Central South University (No. 2020-S053).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Deng X, Wu JB. Research progress on the relationship between microRNAs and liver cancer. Chinese Journal of Digestive Surgery 2015;14:431-3.
- KC M. Steer CJ. Novel mechanisms of chemoresistance by Fusobacterium nucleatum involve not so novel pathways of microRNAs and autophagy. Transl Cancer Res 2018;7:S10-S15. [Crossref]
- Chu Y, Jiang M, Du F, et al. miR-204-5p suppresses hepatocellular cancer proliferation by regulating homeoprotein SIX1 expression. FEBS Open Bio 2018;8:189-200. [Crossref] [PubMed]
- Ye Y, Zhuang J, Wang G, et al. MicroRNA-495 suppresses cell proliferation and invasion of hepatocellular carcinoma by directly targeting insulin-like growth factor receptor-1. Exp Ther Med 2018;15:1150-8. [PubMed]
- Lim JA, Meena NK, Raben N. Pros and cons of different ways to address dysfunctional autophagy in Pompe disease. Ann Transl Med 2019;7:279. [Crossref] [PubMed]
- Zhang Y, Wei Y, Li X, et al. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis 2018;9:130. [Crossref] [PubMed]
- Liu L, Liao JZ, He XX, et al. The role of autophagy in hepatocellular carcinoma: friend or foe. Oncotarget 2017;8:57707-22. [Crossref] [PubMed]
- Li J, Wu PW, Zhou Y, et al. Rage induces hepatocellular carcinoma proliferation and sorafenib resistance by modulating autophagy. Cell Death Dis 2018;9:225. [Crossref] [PubMed]
- Hu P, Cheng B, He Y, et al. Autophagy suppresses proliferation of HepG2 cells via inhibiting glypican-3/wnt/β-catenin signaling. Onco Targets Ther 2018;11:193-200. [Crossref] [PubMed]
- Fan Q, Yang L, Zhang X, et al. Autophagy promotes metastasis and glycolysis by upregulating MCT1 expression and Wnt/β-catenin signaling pathway activation in hepatocellular carcinoma cells. J Exp Clin Cancer Res 2018;37:9. [Crossref] [PubMed]
- Wesselborg S, Stork B. Autophagy signal transduction by ATG proteins: from hierarchies to networks. Cell Mol Life Sci 2015;72:4721-57. [Crossref] [PubMed]
- Fu XT, Shi YH, Zhou J, et al. MicroRNA-30a suppresses autophagy-mediated anoikis resistance and metastasis in hepatocellular carcinoma. Cancer Lett 2018;412:108-17. [Crossref] [PubMed]
- Lin CW, Lin CC, Lee PH, et al. The autophagy marker LC3 strongly predicts immediate mortality after surgical resection for hepatocellular carcinoma. Oncotarget 2017;8:91902-13. [Crossref] [PubMed]
- Chen S, Zhang J, Lin Y. Effects of microRNA-141 on proliferation, apoptosis and cell cycle of liver cancer cells. Chinese Journal of Digestive Surgery 2017;23:401-5.
- Xue H, Tian GY. MiR-429 regulates the metastasis and EMT of HCC cells through targeting RAB23. Arch Biochem Biophys 2018;637:48-55. [Crossref] [PubMed]
- Tao R, Sun WY, Yu DH, et al. Sodium cantharidinate induces HepG2 cell apoptosis through LC3 autophagy pathway. Oncol Rep 2017;38:1233-9. [Crossref] [PubMed]
- Huang KT, Kuo IY, Tsai MC, et al. Factor VII-Induced MicroRNA-135a Inhibits Autophagy and Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Mol Ther Nucleic Acids 2017;9:274-83. [Crossref] [PubMed]
- Xu FF, Xie WF, Zha GQ, et al. MiR-520f promotes cell aggressiveness by regulating fibroblast growth factor 16 in hepatocellular carcinoma. Oncotarget 2017;8:109546-58. [Crossref] [PubMed]
- Hu C, Cui S, Zheng J, et al. MiR-875-5p inhibits hepatocellular carcinoma cell proliferation and migration by repressing astrocyte elevated gene-1 (AEG-1) expression. Transl Cancer Res 2018;7:158-69. [Crossref]
- Wang P, Xie AW, Fan QQ, et al. Mechanism of miR-139-5p targeting transforming growth factor-β1 to inhibit invasion and metastasis of liver cancer cells. Chinese Journal of Digestive Surgery 2016;22:17-23.
- Wang Y, Wang Q, Song J. Inhibition of autophagy potentiates the proliferation inhibition activity of microRNA-7 in human hepatocellular carcinoma cells. Oncol Lett 2017;14:3566-72. [Crossref] [PubMed]
- Boland P, Wu J. Systemic therapy for hepatocellular carcinoma: beyond sorafenib. Chin Clin Oncol 2018;7:50. [Crossref] [PubMed]
- Fang Y, Xue JL, Shen Q, et al. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012;55:1852-62. [Crossref] [PubMed]
- Dai H, Kang DH, Kang B, et al. Effects of MicroRNA-30a-5p on proliferation, apoptosis, invasion and metastasis of hepatoma cell line SMMC-7721. Chinese Journal of Digestive Surgery 2014;22:915-20.
- Ge YY, Shi Q, Zheng ZY, et al. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget 2014;5:6218-28. [Crossref] [PubMed]
- Stiuso P, Potenza N, Lombardi A, et al. MicroRNA-423-5p Promotes Autophagy in Cancer Cells and Is Increased in Serum From Hepatocarcinoma Patients Treated With Sorafenib. Mol Ther Nucleic Acids 2015;4:e233. [Crossref] [PubMed]
- Zhou L, Liu S, Han M, et al. MicroRNA-185 induces potent autophagy via AKT signaling in hepatocellular carcinoma. Tumour Biol 2017;39:1010428317694313. [Crossref] [PubMed]
- Lee YJ, Jang BK. The Role of Autophagy in Hepatocellular Carcinoma. Int J Mol Sci 2015;16:26629-43. [Crossref] [PubMed]
- Chang Y, Yan W, He X, et al. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 2012;143:177-87.e8. [Crossref] [PubMed]
- Wang C, Wang X, Su Z, et al. The novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growth. Oncol Rep 2015;34:1708-16. [Crossref] [PubMed]


