
Overcoming the challenges of imputation of rare variants in a Taiwanese cohort
Significant findings from genome-wide association studies (GWASs) only explain a limited proportion of the genetic variance (i.e., heritability) for many phenotypes (1,2). This proportion potentially increases when all variants typed or tagged by GWAS arrays are considered (3); however, the common disease/common variant (CDCV) hypothesis leads to detection of loci that explain, almost invariably, only a small part of the “missing heritability” (4). The focus of genetic association studies has thus shifted toward rarer alleles with larger effect sizes (5). Researchers refer to variants with minor allele frequencies (MAFs) between 1% and 5% as “less common” variants and those with MAFs <1% as “rare” variants. Rare variants are one of the sources contributing to missing heritability (6), although due to their rarity they may contribute very little to heritability, individually (3). This could be because either the variants are too rare to contribute population-wide, or because the effect size of each variant is, in general, very small. It has been theorized that the severity of diseases in individuals who have the risk variants and display disease symptoms may be largely due to the cumulative effect of a collection of possibly hundreds, or even thousands, of similar conditions that are associated with rare variants at individual loci (7).
Rare variants constitute the bulk of genetic variation in the human genome and are predicted to have larger phenotypic effects than common variants (8); however, it has been challenging to analyze such variants with adequate power in population-based studies due to their poor representation in the genotyping arrays that are typically used in GWASs. The total number of loci that may contribute to a disease’s prevalence is dependent on the disease incidence, the frequency of rare variants per locus, and their effect size (the genotype relative risk) (9). For a disease with high heritability, with the increase of the number of contributing rare alleles in an individual, the relative risk rises steeply under a multiplicative model. However, if each of these variants explains most of the risk in just a few people, their effects will not explain enough of the variance in a total population, and therefore standard GWAS procedures would fail to detect them (9). Furthermore, they are scarcely tagged as single-nucleotide polymorphisms (SNPs) in genome-wide arrays, with the exception of family-based studies and studies with very large sample sizes. Moreover, most CDCV-based GWASs exclude rare variants from their analysis in the early quality control steps. Besides, assaying rare variants directly via arrays is expensive. Hence, researchers opt for the more economical approach of utilizing existing sequencing data as reference panels in order to impute rare variants into existing large datasets for the purpose of characterizing the disease burden of rare variants.
It has been established that the ability to impute a variant accurately is dependent both on the choice of array and the total number of individuals genotyped in the reference panel carrying that variant (10). Simulation studies have found that increasing sample size in the reference panel may improve imputation accuracy, especially for SNPs with relatively low MAFs (11). A large reference panel may capture many less common and rare variants, which should provide a better resolution to establish the haplotype background for observed variants (12). Accordingly, numerous international collaborative projects have made large-scale efforts, over the past decade, to set up successively larger and more genetically diverse resources. The most popular amongst them are the International HapMap Project (13), which produced a reference panel of 420 haplotypes with 3.1 million SNPs in three continental populations; and the 1000 Genomes Project (14,15), which utilized low-coverage whole-genome sequencing to create a reference panel of 5,008 haplotypes with over 88 million variants from 26 worldwide populations. The research community has utilized these primary sources extensively for un-typed SNPs in genomic array datasets. The large number of haplotypes, SNPs, and populations has indeed led to improved genotype imputation accuracy (15), allowing the possibility of imputation and association testing for rare variants as well.
Another panel that has been recently released is the Haplotype Reference Consortium (HRC) panel, which combines datasets from 20 different studies, the majority of which have low-coverage whole-genome sequencing data (4–8× coverage) and are known to consist of samples with predominantly European ancestry (16). The 1000 Genomes Project Phase 3 cohort is a part of this panel, with 64,976 haplotypes at 39,235,157 SNPs having evidence of a minor allele count ≥5. However, it remains controversial as to how effective such approaches are for successfully imputing rare variants, as well as the minimum allele frequency that is accessible by utilizing such reference panels for phasing and imputation.
It has been claimed by some studies that it is unattainable to impute rare variants with MAF <0.03 (17), whereas some other studies have indicated that it is possible to impute not only “less common” variants, but even rare and “very rare” (MAF <0.01) variants using GWAS data (18,19). For instance, one study reported that the HOXB13 G84E mutation (a putative marker for prostate cancer) could not be imputed using the SNPs included on a custom Illumina Collaborative Oncological Gene-Environment Study (iCOGS) array, even though experimental evidence displayed a synthetic association of many common genetic variants in the region with G84E (20). To overcome this bottleneck, a hybrid imputation approach can subsequently be applied to a large cohort of individuals in order to successfully impute such rare mutations. The fundamental idea behind such hybrid approaches is to increase the sample size of the genotype data to identify disease-associated SNPs with higher power, and then use those SNPs to construct a mutation carrier-enriched reference panel. Thereafter, imputation of rare mutations can successfully be achieved through the following consecutive steps: (I) meta-analysis of multiple GWAS studies for a disease of interest, where the genotype data from the GWASs are imputed, (II) combined test of association with the disease under study, over all SNPs, (III) construction of a panel of SNPs that showed evidence of association, and (IV) use of this SNP panel to conduct imputation, thereby increasing the frequency of the putative variants in the populations. Some studies have shown that combining reference panels may increase the number and accuracy of imputed rare variants in comparison to when they are imputed using single reference panels (21). Hence, a customized SNP panel along with other reference panels, such as 1000 Genomes Phase 3, could be combined to identify additional rare SNPs by imputation.
Replicating the G84E mutation from HOXB13 in populations has also been difficult because of its rarity and selectivity for European genomes (carrier frequency of 0.0034 in participants of European ancestry in the 1000 Genomes Project) (20). However, other variants of HOXB13 have been identified in individuals of non-European descent, including G135E in Chinese men, and F127C and G132E in Japanese men (22). This is explained through allelic heterogeneity of genes depending on the population being investigated. Using a reference panel from multiple ethnic groups for SNPs that are not population-specific may still be appropriate to provide accurate imputation (21). However, combining a global reference genome, such as 1000 Genomes [using only population specific samples, such as only East Asian (EAS) or only European (EUR)], with that of a respective ethnically specific one, to conduct imputation, could lead to about 40% more imputed variants than when using 1000 Genomes only (using all ethnicities). The improvement of imputation accuracy is attributed to the fact that such a strategy successfully captures the linkage disequilibrium patterns of the ethnic specific variants which other ethnic groups with different ancestral genetic background, might fail to capture (21). Ethnic composition is an important predictor of imputation accuracy (23). Many studies have validated the accuracy and reliability of imputation of rare variants (24,25), but the focus of most of these studies has been on populations of European descent (25,26). When a publicly available database that was constructed across varied ethnic populations, was used as a reference to conduct imputation, it was found that Europeans displayed the highest accuracy and Africans the lowest (24). Due to the fact that Asian populations possess some unique genetic characteristics, it is neither possible nor appropriate to directly adapt genetic information from studies that have been conducted for Caucasian populations (27). Even large reference panels such as HRC have been shown to display limitations for Han Chinese populations (28), suggesting the necessity of building population-specific reference panels. Factors such as genetic alterations and/or mutations coupled with family history and race have been thought to play important roles in the heritability of genetically complex diseases. Moreover, a higher false positive rate has been observed in imputation from global reference panels compared to imputation performed using a local panel (29). Little research has been conducted in the area of rare variant imputation across ethnic populations, and none in Taiwanese populations. There is no representation of the Taiwanese population in large reference panels such as 1000 Genomes and HRC. The pan-Asian SNP genotyping database (PanSNPdb) (30), which collected SNPs and copy number variations from 1,719 samples in 71 populations including mainland China, India, Indonesia, Japan, Malaysia, the Philippines, Singapore, South Korea, Taiwan, and Thailand, also has a low Taiwanese representation. Therefore, constructing reference panels’ specific for the Taiwanese population is an immediate requirement for conducting rare variant association studies in Taiwanese patients.
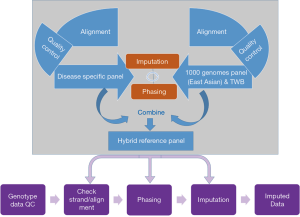
Taiwan has one of the most complete health-related databases in the world, with records covering up to 99% of the population of 23.5 million people (31). Taiwan Biobank (TWB) (https://www.twbiobank.org.tw/new_web_en/index.php), a national database created under the supervision of the Ministry of Health and Welfare, aims to collect data from 200,000 healthy participants and 100,000 individuals with 10 to 15 specific diseases in Taiwan. Utilizing this population-based resource to construct reference panels would enable successful rare variant imputation for the Taiwanese population. The large sample size of the TWB would provide genetic similarity between the reference panel (TWB reference panel) and the target samples, thereby enhancing the accuracy of genotype imputation (32). Specific reference panels that incorporate disease-specific haplotypes can improve imputation of disease-relevant variants, even with the addition of only 100 disease-carrying individuals, as well as the sensitivity and coverage of detected variants in disease-relevant regions. Using this TWB reference panel as the reference genome to conduct meta-analysis would lead to identification of Taiwanese-specific disease variants, which can be used later to customize mutation-enriched panels (disease specific panel). Such customized panels, when coupled with EAS sequences from the 1000 Genomes Phase 3 reference panel and the TWB reference panel, would lead to construction of hybrid reference panels (Figure 1). Using such hybrid panels will (I) provide genetic similarity with Taiwanese individuals for conducting imputation, (II) increase the number of disease-specific haplotypes, and (III) increase the sample size of the reference population, thereby potentially allowing rare variant imputation with enhanced accuracy for Taiwanese cohorts.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research. The article did not undergo external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2395). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature 2009;461:747-53. [Crossref] [PubMed]
- Juang JM, Lu TP, Lai LC, et al. Disease-targeted sequencing of ion channel genes identifies de novo mutations in patients with non-familial Brugada syndrome. Sci Rep 2014;4:6733. [Crossref] [PubMed]
- Hoffmann TJ, Witte JS. Strategies for imputing and analyzing rare variants in association studies. Trends Genet 2015;31:556-63. [Crossref] [PubMed]
- Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 2010;11:446-50. [Crossref] [PubMed]
- Zuk O, Schaffner SF, Samocha K, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A 2014;111:E455-E464. [Crossref] [PubMed]
- Frazer KA, Murray SS, Schork NJ, et al. Human genetic variation and its contribution to complex traits. Nat Rev Genet 2009;10:241-51. [Crossref] [PubMed]
- McClellan JM, Susser E, King MC. Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry 2007;190:194-9. [Crossref] [PubMed]
- Price AL, Kryukov GV, de Bakker PI, et al. Pooled association tests for rare variants in exon-resequencing studies. Am J Hum Genet 2010;86:832-8. [Crossref] [PubMed]
- Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet 2012;13:135-45. [Crossref] [PubMed]
- Sariya S, Lee JH, Mayeux R, et al. Rare variants imputation in admixed populations: Comparison across reference panels and bioinformatics tools. Front Genet 2019;10:239. [Crossref] [PubMed]
- Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457-70. [Crossref] [PubMed]
- Mechanic LE, Chen HS, Amos CI, et al. Next generation analytic tools for large scale genetic epidemiology studies of complex diseases. Genet Epidemiol 2012;36:22-35. [Crossref] [PubMed]
- Consortium IH. The international HapMap project. Nature 2003;426:789. [Crossref] [PubMed]
- Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015;526:75-81. [Crossref] [PubMed]
- Shi S, Yuan N, Yang M, et al. Comprehensive assessment of genotype imputation performance. Hum Hered 2018;83:107-16. [Crossref] [PubMed]
- McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279-83. [Crossref] [PubMed]
- Zheng HF, Ladouceur M, Greenwood CM, et al. Effect of genome-wide genotyping and reference panels on rare variants imputation. J Genet Genomics 2012;39:545-50. [Crossref] [PubMed]
- Joshi PK, Prendergast J, Fraser RM, et al. Local exome sequences facilitate imputation of less common variants and increase power of genome wide association studies. PLoS One 2013;8:e68604. [Crossref] [PubMed]
- Holm H, Gudbjartsson DF, Sulem P, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet 2011;43:316-20. [Crossref] [PubMed]
- Saunders EJ, Dadaev T, Leongamornlert DA, et al. Fine-mapping the HOXB region detects common variants tagging a rare coding allele: evidence for synthetic association in prostate cancer. PLoS Genet 2014;10:e1004129. [Crossref] [PubMed]
- Chou WC, Zheng HF, Cheng CH, et al. A combined reference panel from the 1000 Genomes and UK10K projects improved rare variant imputation in European and Chinese samples. Sci Rep 2016;6:39313. [Crossref] [PubMed]
- Hayano T, Matsui H, Nakaoka H, et al. Germline variants of prostate cancer in Japanese families. PLoS One 2016;11:e0164233. [Crossref] [PubMed]
- Lee D, Bigdeli TB, Williamson VS, et al. DISTMIX: direct imputation of summary statistics for unmeasured SNPs from mixed ethnicity cohorts. Bioinformatics 2015;31:3099-104. [Crossref] [PubMed]
- Huang L, Li Y, Singleton AB, et al. Genotype-imputation accuracy across worldwide human populations. Am J Hum Genet 2009;84:235-50. [Crossref] [PubMed]
- Zhao Z, Timofeev N, Hartley SW, et al. Imputation of missing genotypes: an empirical evaluation of IMPUTE. BMC Genet 2008;9:85. [Crossref] [PubMed]
- Nothnagel M, Ellinghaus D, Schreiber S, et al. A comprehensive evaluation of SNP genotype imputation. Hum Genet 2009;125:163-71. [Crossref] [PubMed]
- Pillai NE, Okada Y, Saw WY, et al. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum Mol Genet 2014;23:4443-51. [Crossref] [PubMed]
- Lin Y, Liu L, Yang S, et al. Genotype imputation for Han Chinese population using Haplotype Reference Consortium as reference. Hum Genet 2018;137:431-6. [Crossref] [PubMed]
- Surakka I, Sarin AP, Ruotsalainen SE, et al. The rate of false polymorphisms introduced when imputing genotypes from global imputation panels. BioRxiv 2016. doi: https://doi.org/
10.1101/080770 . - Ngamphiw C, Assawamakin A, Xu S, et al. PanSNPdb: the Pan-Asian SNP genotyping database. PLoS One 2011;6:e21451. [Crossref] [PubMed]
- Lin JC, Fan CT, Liao CC, et al. Taiwan Biobank: making cross-database convergence possible in the Big Data era. Gigascience 2018;7:1-4. [Crossref] [PubMed]
- Schurz H, Müller SJ, Van Helden PD, et al. Evaluating the accuracy of imputation methods in a five-way admixed population. Front Genet 2019;10:34. [Crossref] [PubMed]


