
Bioinformatics analysis to reveal key biomarkers for early and late hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is a usual malignant tumor with an increasing incidence and one of the leading causes of cancer-associated deaths globally (1,2). Viral infections are often associated with liver cirrhosis and HCC (3). The therapeutic strategies of HCC depend on disease stage, operative tolerance, and possibility of liver transplant (4). HCC remains a challenging clinical problem with high incidence, limited therapeutic strategies, and poor prognosis. Therefore, it is a pressing task to screen potential biomarkers and uncover the detailed molecular mechanisms underlying of HCC.
With the advent of transcriptome analysis, bioinformatics have become most common ways to identify potential biomarkers of various diseases (5,6). Increasing evidences have indicated that lncRNAs plays a key roles in tumor-related biological processes, such as proliferation, invasion and metastasis (7,8). Recently, many lncRNAs are considered to be novel potential biomarkers and therapeutic targets for various cancers (9,10). The emerging roles of lncRNAs in HCC have also been explored in previous studies (11-13). However, there are few studies on biomarkers of lncRNA in early and late HCC.
In the current study, we identify shared differentially expressed lncRNAs (DElncRNAs) and mRNAs (DEmRNAs) between early and late HCC and normal tissue base on the Cancer Genome Atlas (TCGA) data. For the shared DEmRNAs, functional annotation and protein-protein interaction network construction were performed. Furthermore, DElncRNAs-DEmRNAs co-expression network of early and late HCC were performed. The expression levels of candidate genes were verified using quantitative real time polymerase chain reaction (qRT-PCR). Our study identified potential key DEmRNAs and DElncRNAs in early and late HCC, and provided novel insights into the mechanisms of lncRNAs in HCC.
Methods
Data collection
The lncRNA and mRNA gene expression profiles and corresponding clinical data of HCC were download from TCGA (http://tcga data.nci.nih.gov/). Here, 257 early HCC tissues, 65 late HCC tissues and 50 normal adjacent samples from patients with HCC were enrolled.
Differential expression analysis
The undetectable lncRNAs and mRNAs (with read count value =0 in more than 20% HCC case) were filtered and deleted. The DElncRNAs and DEmRNAs in HCC compared to adjacent normal tissues were identified using the R package DESeq2. The thresholds for differential expression analysis were defined as P value <0.01 and |Log2fold change|>1.5. We used the R package v3.3.3 (https://www.r-project.org/) to perform hierarchical clustering analysis of DElncRNAs and DEmRNAs.
Functional annotation
To discover the biological functions and the potential pathways of DEmRNAs in HCC, Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were retrieved using DAVID 6.8 (https://david.ncifcrf.gov/). The terms with P value < 0.05 were significant results.
Protein-protein interaction network construction
The protein-protein interaction network was predicted using the Biological General Repository for Interaction Datasets (http://thebiogrid.org/), and then the protein-protein interaction network was presented by the Cytoscape (3.7.1) (http://www.cytoscape.org/). The nodes represent proteins and edges connect the nodes to show their relationship.
DEmRNA-DElncRNA interaction analysis
The correlation between the lncRNAs and DEmRNAs were analyzed by the Pearson correlation coefficient. The threshold for DElncRNA-DEmRNA co-expression pairs was P value <0.05 and |r|>0.80. We used the Cytoscape (3.7.1) to build the DElncRNA-DEmRNA co-expression network.
qRT-PCR verification
Three gene (CENPF, KIFC1 and ECM1) and three lncRNAs (CTD-2510F5.4, AC092171.4 and HAND2-AS1) were selected as candidate genes for qRT-PCR verification. Here, 6 HCC patients were included in our study from the Department of Hepatobiliary Surgery, The Third Hospital of Hebei Medical University. Twelve tissue samples including 6 HCC tumor samples and 6 cancer adjacent samples were obtained. This study was approved by the institutional ethics committee of The Third Hospital of Hebei Medical University (No. 2018-025-1) and conducted in accordance with the Declaration of Helsinki. The signed informed consents of all the participants were obtained.
Total RNA was isolated using TRizol reagent kit (Invitrogen, China). SuperScript III Reverse Transcription Kit (Invitrogen, China) was utilized to obtain the complementary DNA. With Super Real PreMix Plus SYBR Green (Invitrogen, China), quantitative real-time PCR were generated using the ABI 7500 system (Applied Biosystems, USA). ACTB and GADPH acted as the internal criterion for the targeted genes. The 2−ΔΔCt method was used to address the data.
Results
Identification of DEmRNAs and DElncRNAs
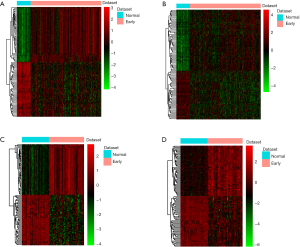
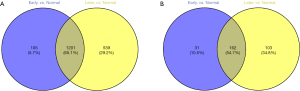
After data processing, we screened 1,306 DEmRNAs (934 up-regulated DEmRNAs and 372 down-regulated DEmRNAs) and 193 DElncRNAs (149 up-regulated DElncRNAs and 44 down-regulated DElncRNAs) in early HCC samples compared with normal samples. A total of 1,740 DEmRNAs (1,222 up-regulated DEmRNAs and 518 down-regulated DEmRNAs) and 265 DElncRNAs (192 up-regulated DElncRNAs and 73 down-regulated DElncRNAs) were obtained between late HCC and normal samples. Figure 1A,B demonstrated hierarchical clustering analysis of the top 100 DEmRNAs and DElncRNAs between early HCC and normal tissue, respectively. Figure 1C,D showed hierarchical clustering analysis of the top 100 DEmRNAs and DElncRNAs between late HCC and normal tissue, respectively. Moreover, a total of 1,201 shared DEmRNAs and 162 shared DElncRNAs were identified in both early and late HCC compared with normal controls. Figure 2 was Venn diagram of DEmRNAs and DElncRNAs in early and late HCC.
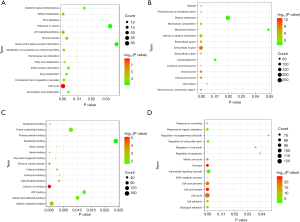
Functional annotation of shared DEmRNAs in early and late HCC
A total of 1,201 shared DEmRNAs in early and late HCC were used to perform the the GO and KEGG pathway enrichment analysis (Figure 3). GO enrichment analysis displayed that M phaseon (P=8.55E-25), cell cycle phasee (P=2.01E-24), chromosome, centromeric region (P=7.50E-13) and calcium ion binding (P=8.65E-06) were significantly enriched GO terms. KEEG enrichment analysis displayed that cell cycle (P=1.92E-09), p53 signaling pathway (P=7.50E-06), Retinol metabolism (P=4.20E-05) and metabolism of xenobiotics by cytochrome P450 (P=5.35E-04) were four significantly enriched pathways.
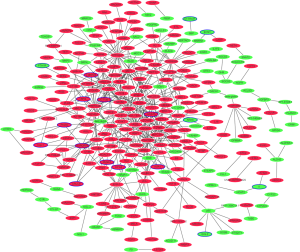
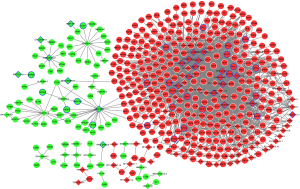
Protein-protein interaction network of shared DEmRNAs in early and late HCC
The protein-protein interaction network was consisted of 292 nodes and 504 edges (Figure 4). Top 10 proteins with higher degrees, including CDK1 (degree=35), AURKA (degree=29), CDC20 (degree=26), PLK1 (degree=23), AURKB (degree=21), HIST1H2BG (degree=17), BUB1B (degree=17), CCNA2 (degree=17), CCNB1 (degree=16) and CDT1 (degree=15).
DElncRNAs-DE mRNAs co-expression network of early HCC
A total of 87 DElncRNA-DEmRNA co-expression pairs including 18 DElncRNAs and 85 DEmRNAs were identified with absolute value of P value <0.05 and |r|>0.80. The co expressed DElncRNA-DEmRNA network (Figure 5) was consisted of 103 nodes and 87 edges, and its hub lncRNAs were CTD-2510F5.4 (degree=58) and HAND2-AS1 (degree=11).
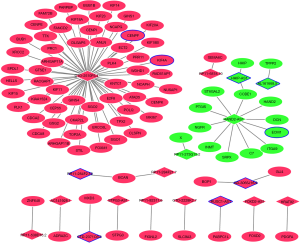
DElncRNAs-DEmRNAs co-expression network of late HCC
A total of 2,339 DElncRNA-DEmRNA co-expression pairs including 92 DElncRNAs and 389 DEmRNAs were identified with absolute value of P value <0.05 and |r|>0.80. The co-expressed DElncRNA-DEmRNA network (Figure 6) was consisted of 481 nodes and 2,339 edges, and its hub lncRNAs were CTD-2510F5.4 (degree=225), RP11-196G18.22 (degree=149), SNHG1 (degree=146), RP5-821D11.7 (degree=146), TMPO-AS1 (degree=144) and HAND2-AS1 (degree=14).
qRT-PCR verification
As Figure 7 displayed, CTD-2510F5.4, HAND2-AS1 and ECM1 were down-regulated and AC092171.4, CENPF and KIFC1 were up-regulated in HCC compared to adjacent tissues. Base on the TCGA results, HAND2-AS1 and ECM1 were down-regulated while the other lncRNAs and DEmRNAs were up-regulated in HCC compared to adjacent tissues. In generally, the confirmation results of qRT-PCR were consistent with our integrated analysis.

Discussion
HCC is a prevalent malignancy that is considered the leading cause of cancer-related deaths worldwide due to the lack of effective treatment (3). Thence, it is urgent to discover potential biomarkers and therapeutic targets for HCC. In this study, a total of 1201 shared DEmRNAs and 162 shared DElncRNAs were identified in both early and late HCC compared with normal controls. For the shared DEmRNAs, functional annotation and protein-protein interaction network construction were performed. Among which, CDK1 (degree=35) was key nodes. In addition, the expressions of 6 candidate genes were verified by the qRT-PCR experiments.
Cyclin dependent kinase 1 (CDK1), belongs to the Ser/Thr protein kinase family, is associated with cell cycle. A recent study has been reported that metformin inhibits the proliferation of HCC cells through inducing G2/M arrest and reduces the expression of CDK1 (14). Wu et al. found that high expression levels of CDK1 in HCC were associated with poor overall survival (15). CDK1 might be involved in progression of HCC via interaction with PLK1 and FOS (16). In this study, CDK1 was also up-regulated in HCC and significantly enriched in pathways of p53 signaling pathway. Paired-related homeobox 1 involves in metastasis and apoptosis of HCC cells by regulating p53-dependent signaling pathway (17). Therefore, we speculated that CDK1 might play a role in pathogenesis of HCC by regulating p53-dependent signaling pathway.
Kinesin family member C1 (KIFC1), a member of the kinesin superfamily, is to act as a motor protein and move towards the minus-end of the microtubule to further affect its dynamics and functions (18). KIFC1 regulates the formation of centrosomes, and it has been found that upregulation of KIFC1 results in the formation of unipolar spindles in cancer cells (19). It has been reported that KIFC1 is a potential marker that predicts an aggressive disease course in ovarian cancer (19). KIFC1 is overexpressed in HCC and that its expression is closely related to the progression and metastasis of HCC (20). High expression of KIFC1 is associated with the progression of HCC, indicating that KIFC1 is a putative marker for prognostic and therapeutic in HCC (21). In this study, we found that KIFC1 was up-regulated in both TCGA integration analysis and qRT-PCR validation, indicating that it might be a potential therapeutic target for HCC.
Centromere protein F (CENPF) is a cell cycle-associated nuclear antigen associated with the centromere-kinetochore complex whose expression and subcellular localization depend on the cell cycle and degrade rapidly at the end of cell division (22). CENPF is defined as a biomarker of cell proliferation in various malignant tumors, including breast cancer (23), HCC (22), and other tumors (24). Researchers have reported that CENPF may be a biomarkers in diagnosis and prognosis of HCC (22,25). In the present study, we found that CENPF was also up-regulated in both TCGA integration analysis and qRT-PCR validation. CENPF was co-expressed with CTD-2510F5.4 in both early and late HCC. CTD-2510F5.4 expression is positive correlation with prognosis in lung adenocarcinoma (26). CTD-2510F5.4 is a malignant phenotype associated lncRNAs and a biomarker for the diagnosis and prognosis of gastric cancer (27). Therefore, we hypothesized that CTD-2510F5.4 might be involved in progression of HCC by regulating CENPF.
Extracellular matrix protein 1 (ECM1), an 85 kDa glycoprotein, is involved in a number of biological processes including mineralization, cell proliferation and angiogenesis. ECM1 modulates the epithelial–mesenchymal transition progression and cell proliferation of breast cancer via regulating the association between β-catenin and the MUC1 (28). Chen et al. found that ECM1 accelerates migration and invasion of HCC via mediating epithelial-mesenchymal transition (29). In this study, we found that ECM1 was down-regulated in both TCGA integration analysis and qRT-PCR validation, and ECM1 was co-expressed with HAND2-AS1 in both early and late HCC. A study has suggested that HAND2-AS1 is down-regulated in HCC tissues, and HAND2-AS1 is associated with invasion of HCC cells (30). Here, HAND2-AS1 was reduced in HCC, which provided further evidence for previous study. Therefore, we speculated that HAND2-AS1 might be involved with the pathogenesis of HCC through regulating ECM1.
In summary, a total of 1,201 shared DEmRNAs and 162 shared DElncRNAs were identified in both early and late HCC compared with normal controls. Among which, CDK1, KIFC1, CENPF, ECM1, CTD-2510F5.4 and HAND2-AS1 were identified as potential biomarkers for HCC. Overall, the confirmation results of qRT-PCR were generally consistent with our integrated analysis. Several limitations of our study should be mentioned. More samples are needed to validate expression of DEmRNAs and DElncRNAs. Besides, in vivo and in vitro experiments are necessary to reveal biological functions of DEmRNAs and DElncRNAs in HCC in future study.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2238). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional ethics committee of The Third Hospital of Hebei Medical University (No. 2018-025-1) and conducted in accordance with the Declaration of Helsinki (as revised in 2013). The signed informed consents of all the participants were obtained.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rawla P, Sunkara T, Muralidharan P, et al. Update in global trends and aetiology of hepatocellular carcinoma. Contemp Oncol (Pozn) 2018;22:141-50. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016;2:16018. [Crossref] [PubMed]
- Kim Y, Park YH, Hwang S, et al. Diagnostic Role of Blood Tumor Markers in Predicting Hepatocellular Carcinoma in Liver Cirrhosis Patients Undergoing Liver Transplantation. Ann Transplant 2016;21:660-7. [Crossref] [PubMed]
- Wang Y, Wu N, Liu J, et al. FusionCancer: a database of cancer fusion genes derived from RNA-seq data. Diagn Pathol 2015;10:131. [Crossref] [PubMed]
- Yang H, Li H. CD36 identified by weighted gene co-expression network analysis as a hub candidate gene in lupus nephritis. PeerJ 2019;7:e7722. [Crossref] [PubMed]
- Chen X, Tang FR, Arfuso F, et al. The Emerging Role of Long Non-Coding RNAs in the Metastasis of Hepatocellular Carcinoma. Biomolecules 2019;10:66. [Crossref] [PubMed]
- Cui Z, Han B, Wang X, et al. Long Non-Coding RNA TTN-AS1 Promotes the Proliferation and Invasion of Colorectal Cancer Cells by Activating miR-497-Mediated PI3K/Akt/mTOR Signaling. Onco Targets Ther 2019;12:11531-9. [Crossref] [PubMed]
- Zeng JH, Liang L, He RQ, et al. Comprehensive investigation of a novel differentially expressed lncRNA expression profile signature to assess the survival of patients with colorectal adenocarcinoma. Oncotarget 2017;8:16811-28. [Crossref] [PubMed]
- Wei C, Liang Q, Li X, et al. Bioinformatics profiling utilized a nine immune-related long noncoding RNA signature as a prognostic target for pancreatic cancer. J Cell Biochem 2019;120:14916-27. [Crossref] [PubMed]
- Long J, Bai Y, Yang X, et al. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int 2019;19:90. [Crossref] [PubMed]
- Wang F, Xie C, Zhao W, et al. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med 2017;17:33-43. [Crossref] [PubMed]
- Sonohara F, Inokawa Y, Hayashi M, et al. Prognostic Value of Long Non-Coding RNA HULC and MALAT1 Following the Curative Resection of Hepatocellular Carcinoma. Sci Rep 2017;7:16142. [Crossref] [PubMed]
- Zhou J, Han S, Qian W, et al. Metformin induces miR-378 to downregulate the CDK1, leading to suppression of cell proliferation in hepatocellular carcinoma. Onco Targets Ther 2018;11:4451-9. [Crossref] [PubMed]
- Wu CX, Wang XQ, Chok SH, et al. Blocking CDK1/PDK1/beta-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics 2018;8:3737-50. [Crossref] [PubMed]
- Gao X, Wang X, Zhang S. Bioinformatics identification of crucial genes and pathways associated with hepatocellular carcinoma. Biosci Rep 2018;38:BSR20181441. [Crossref] [PubMed]
- Fan M, Shen J, Liu H, et al. Downregulation of PRRX1 via the p53-dependent signaling pathway predicts poor prognosis in hepatocellular carcinoma. Oncol Rep 2017;38:1083-90. [Crossref] [PubMed]
- Xiao YX, Shen HQ, She ZY, et al. C-terminal kinesin motor KIFC1 participates in facilitating proper cell division of human seminoma. Oncotarget 2017;8:61373-84. [Crossref] [PubMed]
- Mittal K, Choi DH, Klimov S, et al. A centrosome clustering protein, KIFC1, predicts aggressive disease course in serous ovarian adenocarcinomas. J Ovarian Res 2016;9:17. [Crossref] [PubMed]
- Han J, Wang F, Lan Y, et al. KIFC1 regulated by miR-532-3p promotes epithelial-to-mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene 2019;38:406-20. [Crossref] [PubMed]
- Fu X, Zhu Y, Zheng B, et al. KIFC1, a novel potential prognostic factor and therapeutic target in hepatocellular carcinoma. Int J Oncol 2018;52:1912-22. [PubMed]
- Sun B, Lin G, Ji D, et al. Dysfunction of Sister Chromatids Separation Promotes Progression of Hepatocellular Carcinoma According to Analysis of Gene Expression Profiling. Front Physiol 2018;9:1019. [Crossref] [PubMed]
- Sun J, Huang J, Lan J, et al. Overexpression of CENPF correlates with poor prognosis and tumor bone metastasis in breast cancer. Cancer Cell Int 2019;19:264. [Crossref] [PubMed]
- Shi J, Zhang P, Liu L, et al. Weighted gene coexpression network analysis identifies a new biomarker of CENPF for prediction disease prognosis and progression in nonmuscle invasive bladder cancer. Mol Genet Genomic Med 2019;7:e982. [Crossref] [PubMed]
- Cai C, Wang W, Tu Z. Aberrantly DNA Methylated-Differentially Expressed Genes and Pathways in Hepatocellular Carcinoma. J Cancer 2019;10:355-66. [Crossref] [PubMed]
- Wang X, Li G, Luo Q, et al. Integrated TCGA analysis implicates lncRNA CTB-193M12.5 as a prognostic factor in lung adenocarcinoma. Cancer Cell Int 2018;18:27. [Crossref] [PubMed]
- Wang Z, Qin B. Prognostic and clinicopathological significance of long noncoding RNA CTD-2510F5.4 in gastric cancer. Gastric Cancer 2019;22:692-704. [Crossref] [PubMed]
- Wu Q, Chen D, Luo Q, et al. Extracellular matrix protein 1 recruits moesin to facilitate invadopodia formation and breast cancer metastasis. Cancer Lett 2018;437:44-55. [Crossref] [PubMed]
- Chen H, Jia W, Li J. ECM1 promotes migration and invasion of hepatocellular carcinoma by inducing epithelial-mesenchymal transition. World J Surg Oncol 2016;14:195. [Crossref] [PubMed]
- Yang Y, Chen L, Gu J, et al. Recurrently deregulated lncRNAs in hepatocellular carcinoma. Nat Commun 2017;8:14421. [Crossref] [PubMed]


