Mutational profiling of melanomas in patients from the southeast coast of China
Introduction
Melanoma is the most lethal form of skin cancer. Based on the anatomical site, it can be classified as cutaneous (with or without chronic sun-induced damage), acral, or mucosal (1). Cutaneous melanoma is the predominant subtype in Caucasian populations (2). However, acral and mucosal melanomas are more common in African and Asian populations (3). Among white people, acral melanomas account for approximately 10% of all cases of melanoma, but in Asians, they account for 50–70% (4,5). The treatment for melanoma include surgery, chemotherapy, radiation therapy, targeted therapy and immunotherapy. Early-stage melanomas often be treated with surgery alone, but more advanced cancers usually require other treatments, sometimes it will use more than one type of treatment.
There have been some genomic profiling studies of melanoma in recent decades. The Cancer Genome Atlas (TCGA) described the landscape of genomic alterations in 333 samples with cutaneous melanoma and established a four-type classification based on the pattern of the most prevalent significantly mutated genes (BRAF, RAS, and NF1) (6). Research consistently shows that BRAF mutations are the most common activating mutation in cutaneous melanoma and represent an essential therapeutic option in advanced disease (6). Studies have found that in acral and mucosal melanoma, KIT mutations have a higher frequency than in cutaneous melanoma. Curtin et al. (7) reported that KIT mutations were found in 29 % of melanomas, but the incidence was approximately 11 percent in a Chinese cohort (8), which indicates that the oncogene mutation status that vary between Caucasians and Asians. Other research that studied the genomic profile of acral melanoma in Asian patients found that the RAS gene was frequently mutated (9). Although there have been other genetic studies of melanoma, relatively few studies focus on Chinese patients, and most of them concerned specific gene mutations (8,10-12). Furthermore, a comprehensive comparison of genomic alterations among Chinese cutaneous, acral, and mucosal melanoma was lacking. In this study, by using targeted NGS sequencing methods, we aimed to evaluate the clinicopathological and genetic characteristics in a cohort of 62 patients from the southeast coast of China with melanoma at different anatomical sites.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1871).
Methods
Patients
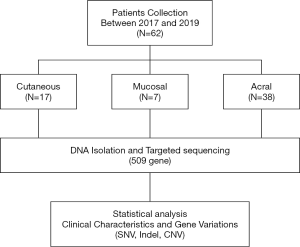
Patients were enrolled from the Zhejiang Cancer Hospital between 2017 and 2019. A total of 62 patients were included in NGS testing. Clinical data were obtained after a comprehensive review of the patients’ medical records. This study was approved by the institutional ethical committee of the medical faculty of Zhejiang Cancer Hospital. All procedures performed in studies involving human participants were in accordance with the institutional research committee and with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all individual participants included in the study.
DNA isolation and targeted sequencing
Tumor tissue was collected, snap-frozen, and stored in liquid nitrogen or embedded in paraffin. Blood samples from the corresponding patient were used as the control. Genomic DNA was extracted from these samples following the protocol described in a previous manuscript (13). Targeted sequencing was performed using multiple gene panels (Genetron Health Co. Ltd., Country), and the Illumina X10 platform was used for sequencing.
Statistical analysis
The sequencing reads were mapped to the human reference genome (hg19) using Burrows-Wheeler Aligner (BWA). Duplicate removal, local realignment, and base quality recalibration were performed using PICARD. Somatic single nucleotide variations (SNVs) were called using Mutect, small indels were called using strelka, and somatic CNVs were called with ExomeCNV. Each variant was confirmed using IGV.
Statistical analyses were conducted using SPSS 23.0. Associations between covariates were investigated using chi-squared tests. A P value of ≤0.05 was interpreted as statistically significant.
Results
Patient clinicopathological characteristics
A total of 62 samples—23 primary tumor and 39 metastases samples—were collected from Zhejiang Cancer Hospital. Seventeen tumors were cutaneous melanoma, 7 were mucosal melanoma, and 38 were acral melanoma (Figure 1). The clinicopathological characteristics of the patients enrolled are summarized in Table 1. In this cohort, 40 (64.5%) patients were male, and 22 (35.5%) were female, and the median age was 62.5 years (ranging from 32 to 85 years). The Breslow thickness, ulceration, sentinel lymph node status was counted among acral, mucosal, and cutaneous melanomas, and no significant differences were found among them (Table 1).
Table 1
| Characteristic | Total | Cutaneous (%) | Mucosal (%) | Acral (%) | P value |
|---|---|---|---|---|---|
| Age (median) | 62.5 | 51 | 63 | 66 | 0.430 |
| Sex | |||||
| Female | 22 | 6 [35] | 4 [57] | 12 [31] | |
| Male | 40 | 11 [65] | 3 [43] | 26 [69] | |
| Neoadjuvant therapies | 0.630 | ||||
| Yes | 12 | 4 [24] | 0 [0] | 8 [21] | |
| No | 47 | 12 [71] | 7 [100] | 28 [74] | |
| Data missing | 3 | 1 [6] | 0 [0] | 2 [5] | |
| Breslow thickness | |||||
| Up to 1 mm | 5 | 2 [12] | 1 [14] | 2 [5] | 0.584 |
| 1.01–2 mm | 9 | 2 [12] | 2 [29] | 5 [13] | |
| 2.01–4 mm | 14 | 5 [29] | 0 [0] | 9 [24] | |
| More than 4 mm | 19 | 4 [24] | 1 [14] | 14 [37] | |
| Data missing | 15 | 4 [24] | 3 [43] | 8 [21] | |
| Ulceration | 0.167 | ||||
| Absent | 38 | 12 [71] | 6 [86] | 20 [53] | |
| Present | 24 | 5 [29] | 1 [14] | 18 [47] | |
| SLN status | 0.699 | ||||
| Negative | 38 | 11 [65] | 3 [43] | 24 [63] | |
| Positive | 22 | 5 [29] | 4 [57] | 13 [34] | |
| Data missing | 2 | 1 [6] | 0 [0] | 1 [3] | |
| TNM stage | 0.214 | ||||
| I | 6 | 3 [18] | 2 [29] | 1 [3] | |
| II | 17 | 4 [24] | 2 [29] | 11 [29] | |
| III | 29 | 9 [53] | 3 [43] | 17 [45] | |
| IV | 7 | 0 [0] | 0 [0] | 7 [18] | |
| Data missing | 3 | 1 [6] | 0 [0] | 2 [5] | |
SLN, sentinel lymph node.
The genetic landscape of melanomas in patients from the southeast coast of China
Targeted sequencing was carried out from 62 patients on paired tumor and normal genomic DNA of the germline, with a mean coverage of 800×. Single nucleotide variants, small indels, fusions, and CNVs were analyzed to evaluate the genomic changes in these patients. In total, 286 aberrations (ranging from 1 to 24 per sample), including 205 missense mutations, 18 frameshift mutations, seven small indels, 16 nonsense mutations, three fusions, six splice site mutations, one translation start site mutation, and 30 amplification sites, were found by targeted sequencing.
The mutational landscape is shown in Figure 2. BRAF was the most frequently mutated gene in this cohort (n=18, 29%); of the samples with BRAF hotspot mutations, 13 (81.2%) had V600E mutation, and the remaining three samples had other mutations (V600K and D594, Figure 3). Additionally, two patients carried BRAF deletions and fusions, which have not been reported in melanoma before and have an unknown effect on gene function. Another recurrent gene family was the RAS family (n=18, 29%); the most frequently mutated isoform was NRAS (n=13, 72.2%), followed by KRAS (n=4, 22.2%) and HRAS (n=1, 5.6%). RAS mutation was mutually exclusive with BRAF gene mutation, except in one sample. Mutation of KIT, also an essential gene in melanoma, was found in six individuals (9.7%), and KIT was not co-mutated with the BRAF or RAS gene. NF1 mutation was detected in five individuals (8.1%), all of whom had a loss of function mutation. CTNNB1 was mutated in 7 samples (11.3%); 6 of these mutations were found in exon three and were activating mutations, and one was in exon ten and had an unknown effect (Table 2).
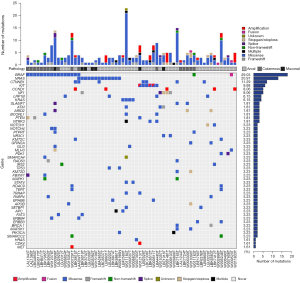
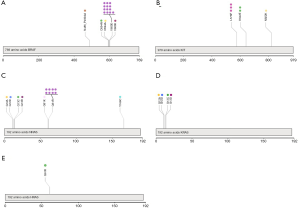
Table 2
| Gene | N (%) | Exon | Amino acid change | Location |
|---|---|---|---|---|
| BRAF | 18 (29) | 15 | p.V600E (n=13, 81.2%) | Acral & cutaneous & mucosal |
| 15 | p.V600K (n=1, 5.6%) | Cutaneous | ||
| 15 | p.D594N (n=1, 5.6%) | Cutaneous | ||
| 15 | p.D594A (n=1, 5.6%) | Acral | ||
| 8 | BRAF-NRF1 (n=1, 5.6%) | Acral | ||
| 12 | p.N486_P490del (n=1, 5.6%) | Cutaneous | ||
| NRAS | 13 (21) | 1 | p.G12A (n=1, 7.7%) | Mucosal |
| 1 | p.G12D (n=1, 7.7%) | Acral | ||
| 1 | p.G13C (n=1, 7.7%) | Mucosal | ||
| 1 | p.G13R (n=1, 7.7%) | Cutaneous | ||
| 2 | p.Q61K (n=1, 7.7%) | Acral | ||
| 2 | p.Q61R (n=8, 61.5%) | Acral & mucosal | ||
| KRAS | 4 (6.5) | 1 | p.G12A (n=1, 25%) | Acral |
| 1 | p.G12D (n=1, 25%) | Acral | ||
| 1 | p.G13C (n=1, 25%) | Acral | ||
| 1 | p.G13D (n=1, 25%) | Acral | ||
| HRAS | 1 (1.6) | 2 | p.Q61R (n=1, 100%) | Acral |
| CTNNB1 | 7 (11.3) | 3 | p.G34V (n=1, 14.3%) | Acral |
| 3 | p.S33C (n=1, 14.3%) | Acral | ||
| 3 | p.S37Y (n=1, 14.3%) | Acral | ||
| 3 | p.D32V (n=1, 14.3%) | Acral | ||
| 3 | p.D32Y (n=1, 14.3%) | Cutaneous | ||
| 3 | p.S33Y (n=1, 14.3%) | Acral | ||
| 10 | p.T510N (n=1, 14.3%) | Acral | ||
| KIT | 6 (9.7) | 11 | p.L576P (n=3, 50%) | Acral |
| 13 | p.K642E (n=2, 33.3%) | Acral & mucosal | ||
| 17 | p.N822K (n=1, 16.7%) | Cutaneous |
Gene mutation profiles of melanomas at different anatomical sites
According to the molecular classification described in an earlier report, cutaneous melanoma can be divided into four subtypes. In our cohort, the most common genomic subtype of cutaneous melanoma was BRAF-mutated (n=12, 70.6%), followed by triple wild type (n=5, 29.4%); no NF1 mutation was found in our cutaneous samples. In the BRAF-mutated group, nine samples with the BRAF V600E mutation, 1 with the V600K mutation, 1 with small deletions, and 1 with the D594N mutation accompanied by the NRAS G13R mutation were found. In the triple wild-type group, four samples had other functional mutations, such as KIT, MYC, PBRM1, and ARID2 mutations. Mutation of TP53, one of the previously reported significantly mutated genes in cutaneous melanoma (2), was not found in our cutaneous melanoma cohort, and only two samples with TP53 mutation were found in the acral group.
The mucosal group had a small sample size (n=7). NRAS had the highest mutation frequency (n=4, 57.1%), BRAF V600E mutation was detected in one sample (14.3%), and the remaining two samples had KIT and NF1 as driver mutations separately. In the acral group (n=38), the most frequent mutation was RAS mutation (n=13, 34.2%); 8 samples had NRAS mutation; 4, KRAS; and 1, HRAS. BRAF mutation was detected in 5 (13.2%) individuals; three of these mutations were V600E mutations. The KIT mutation was found in 4 (10.5%) individuals, and another 3 (7.9%) had NF1 dysfunctional mutations. There were 5 (13.2%) samples with CTNNB1 activation mutations (Table 3).
Table 3
| Anatomical sites | Number | Important gene variation |
|---|---|---|
| Cutaneous | 11 | BRAF |
| 1 | BRAF, NRAS | |
| 1 | KIT | |
| 1 | RBPM1 | |
| 1 | MYC amplification | |
| 1 | ARID2 | |
| Mucosal | 4 | NRAS |
| 1 | BRAF | |
| 1 | KIT | |
| 1 | NF1 | |
| Acral | 7 | NRAS |
| 4 | KRAS | |
| 3 | BRAF | |
| 2 | KIT, CTNNB1 | |
| 2 | KIT | |
| 1 | NRAS, CTNNB1 | |
| 1 | HRAS | |
| 1 | BRAF, CTNNB1 | |
| 1 | BRAF fusion | |
| 1 | CTNNB1 | |
| 3 | NF1 |
Additionally, 13 patients had gene amplification: 10 (26.3%) in the acral group, 1 (14.3%) in the mucosal group, and 2 (11.8%) in the cutaneous group. The most frequently amplified gene was CCND1, consistent with the results of a previous study (2). Also, gene amplification tended to occur in metastasis samples (n=8) rather than primary samples (n=5).
Discussion
In this study, we used NGS sequencing to evaluate samples from 62 melanomas in a Chinese population from the southeast coast of China and determine their genetic profile. The results showed that BRAF, RAS, CTNNB1, KIT, and NF1 were the most recurrently altered genes in our cohort. The mutational profiles were different in the three melanoma types according to the anatomical site and may help us to understand better the genetic cause of melanoma in the Chinese population.
BRAF, NRAS, and KIT are the critical driver mutations in melanoma. In our cohort, RAS mutation was the most often found mutation except for BRAF mutation. RAS mutation usually occurs at codon 12, 13, or 61 and maintains RAS in a constitutively active state (14,15). Si et al. (10) found that the frequency of NRAS gene mutation was 7.2% (31/432) in the Chinese cohort, with mutation rates of 8.8%, 9.2%, and 2.5% in acral, mucosal and cutaneous melanoma, respectively. In our cohort, the total NRAS mutation frequency was 21% (13/62), which was higher than previously reported (16). Also, the frequencies at different anatomical sites were 21% (8/38), 57% (4/7), and 5.9% (1/17) in acral, mucosal, and cutaneous melanoma, respectively. For the difference of our cohort with these two studies (10,16), we found that the method to test mutation for them was PCR followed Sanger sequencing, this method may not be sensitive for the mutation frequency less than 10%. In our cohort, we use NGS to test the mutation with the limit of detection of 1%, and 6 (46%) samples with the mutation frequency of NRAS less than 10% were found. Except that, the small size of our cohort may also be a reason for the high mutation rate of NRAS. Although no drug can target NRAS directly, inhibition of MEK may be useful for treating NRAS mutant melanoma (17). In a phase III trial in NRAS mutant stage IIIC or stage IV melanoma, either untreated or with progression on immunotherapy, the NRAS inhibitor binimetinib showed a response rate of 15%, with a median PFS time of 2.8 months compared with 1.5 months in the control group (HR 0.62, P<0.01) (18). Also, this trial showed that in melanomas with NRAS mutation, binimetinib may be a choice for patients with progressive disease after first-line immunotherapy, especially when there are no other therapeutic options. As the higher mutation rate of NRAS in our cohort, maybe the clinical trial of MEK inhibitor will give these patients more treatment options.
In addition to the significantly mutated genes, other cancer driver genes showed mutations. CTNNB1 mutation that was not previously reported as a recurrently mutated gene was found in 7 patients in our cohort (n=62), 6 of whom were in the acral melanoma cohort. In another study, Newell et al. (19) found that six samples (n=112) in the mucosal melanoma cohort had CTNNB1 mutations. CTNNB1 is an essential gene in the Wnt/β-catenin pathway, and activation of this pathway has been demonstrated to cause T cell exclusion (20). Also, some advanced stage mucosal melanoma patients with CTNNB1 activating mutations may have relatively more reduced responses to immunotherapy. These results suggest that patients with CTNNB1 activating mutations may need to be more cautious when considering immunotherapy.
Also, there were some limitations to our study. First, the sample size was not significant and was a signal center study, the samples still needed to be collecting, and the results will be tested in a multicenter study in the future. Second, this study focuses on the genetic variation, and the relationship of genetic changes with the clinical outcome will still be explored in the next step. In conclusion, our study profiled the mutational landscape of melanoma in patients from the southeast coast of China. In addition to the most frequently mutated genes (BRAF, RAS, KIT) reported in other studies, we found some new recurrent gene mutations, such as CTNNB1 mutation in acral melanoma, which was not reported in other studies. A better understanding of the molecular mechanism of melanoma may aid the design of clinical trials and explain drug efficacy in the future.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1871
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1871
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1871). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the institutional ethical committee of the medical faculty of Zhejiang Cancer Hospital. All procedures performed in studies involving human participants were in accordance with the institutional research committee and with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all individual participants included in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yentz S, Lao CD. Immunotherapy for mucosal melanoma. Ann Transl Med 2019;7:S118. [Crossref] [PubMed]
- Xu TX, Yu SF, Ma M, et al. cMET-N375S germline mutation is associated with poor prognosis of melanoma in Chinese patients. Transl Cancer Res 2018;7:248-56. [Crossref]
- Desai A, Ugorji R, Khachemoune A. Acral melanoma foot lesions. Part 1: epidemiology, aetiology, and molecular pathology. Clin Exp Dermatol 2017;42:845-8. [Crossref] [PubMed]
- Moon KR, Choi YD, Kim JM, et al. Genetic Alterations in Primary Acral Melanoma and Acral Melanocytic Nevus in Korea: Common Mutated Genes Show Distinct Cytomorphological Features. J Invest Dermatol 2018;138:933-45. [Crossref] [PubMed]
- Sheen YS, Liao YH, Lin MH, et al. Insulin-Like Growth Factor II mRNA-Binding Protein 3 Expression Correlates with Poor Prognosis in Acral Lentiginous Melanoma. PLoS One 2016;11:e0147431. [Crossref] [PubMed]
- Cancer Genome Atlas N. Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681-96. [Crossref]
- Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006;24:4340-6. [Crossref] [PubMed]
- Kong Y, Si L, Zhu Y, et al. Large-scale analysis of KIT aberrations in Chinese patients with melanoma. Clin Cancer Res 2011;17:1684-91. [Crossref] [PubMed]
- Sheen YS, Tan KT, Tse KP, et al. Genetic alterations in primary melanoma in Taiwan. Br J Dermatol 2019; [Epub ahead of print]. [PubMed]
- Si L, Kong Y, Xu X, et al. Prevalence of BRAF V600E mutation in Chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer 2012;48:94-100. [Crossref] [PubMed]
- Si L, Guo J. C-kit-mutated melanomas: the Chinese experience. Curr Opin Oncol 2013;25:160-5. [Crossref] [PubMed]
- Bai X, Kong Y, Chi Z, et al. MAPK Pathway and TERT Promoter Gene Mutation Pattern and Its Prognostic Value in Melanoma Patients: A Retrospective Study of 2,793 Cases. Clin Cancer Res 2017;23:6120-7. [Crossref] [PubMed]
- Pan C, Diplas BH, Chen X, et al. Molecular profiling of tumors of the brainstem by sequencing of CSF-derived circulating tumor DNA. Acta Neuropathol 2019;137:297-306. [Crossref] [PubMed]
- Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature 1987;327:293-7. [Crossref] [PubMed]
- Fecher LA, Cummings SD, Keefe MJ, et al. Toward a molecular classification of melanoma. J Clin Oncol 2007;25:1606-20. [Crossref] [PubMed]
- Vicente A, Crovador CS, Macedo G, et al. Mutational Profile of Driver Genes in Brazilian Melanomas. J Glob Oncol 2019;5:1-14. [Crossref] [PubMed]
- Sun J, Carr MJ, Khushalani NI. Principles of Targeted Therapy for Melanoma. Surg Clin North Am 2020;100:175-88. [Crossref] [PubMed]
- Dummer R, Schadendorf D, Ascierto PA, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol 2017;18:435-45. [Crossref] [PubMed]
- Newell F, Kong Y, Wilmott JS, et al. Whole-genome landscape of mucosal melanoma reveals diverse drivers and therapeutic targets. Nat Commun 2019;10:3163. [Crossref] [PubMed]
- Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231-5. [Crossref] [PubMed]