Quantitative accuracy of radiomic features of low-dose 18F-FDG PET imaging
Introduction
A quantitative radiomic approach or radiomics has been shown promising in diagnosis, response prediction and prognostication (1). It is now a very active research in oncological imaging by extracting high dimensional radiomic features (RFs) to characterize tumor. The most commonly used RF are first-order statistics (FOS) and texture features (TFs), including second-order features calculated from gray level co-occurrence matrix (GLCM) and high-order features including the gray-level run length matrix (GLRLM) (2), gray-level size zone matrix (GLSZM) (3), neighborhood gray-level difference matrix (NGLDM) (4) and neighbor gray-tone difference matrix (NGTDM) (5). Maximum standardized uptake value (SUVmax), mean SUV (SUVmean) and peak SUV (SUVpeak) belong to FOS, which are commonly used in the clinic to measure tissue uptake of 18F-FDG without concerning for voxel spatial relationships (6). The utilities of PET based radiomics have been shown in various tumor types [including pancreatic (7), brain (8), lung (9), esophageal (10) and oropharyngeal (11) cancer] mainly for predicting survival. The more comprehensive characterization of PET based texture analysis for clinical applications could be found in the review articles (12,13).
Quantification in PET imaging using SUV measures is subject to many errors from technical, physical and biologic factors, including scan acquisition parameters, image reconstruction parameters, patient motion and breathing, uptake period, partial volume effect and region of interest (ROI) delineation (14). These errors have been directly realized in SUV measures and have effect on PET clinical applications (15). Some errors have been investigated in RF including motion (16-18), reconstruction settings (19,20), partial volume effect (21) and tumor delineation (21,22).
The clinical value of PET/CT scanning is unquestionable. A higher dose of PET results in a better image but with a cost of higher radiation exposure to patients and hospital personnel. Although the linear no-threshold hypothesis is controversial and low radiation such as radiation from medical imaging perhaps helps prevent cancer instead of increasing cancer (23), the public worry the carcinogenic risk due to medical imaging radiation. Low-dose PET/CT imaging is desired for patients, especially for younger patients. The advances of time of flight (TOF) technology and point spread function (PSF) based reconstruction algorithm could possibly increase sensitivity to detect lesions for some specific clinical tasks (24-26). In addition, low-dose PET/CT imaging is desired for pediatric patients or patients requiring repeated scans. To the best of our knowledge, no studies have been done to investigate RF stability under low-dose PET imaging.
The objective of this study was to evaluate the effect of lowering counts on RF in comparison with the “true values” calculated with full count data. We present the following article in accordance with the STROBE reporting checklist (27) (available at http://dx.doi.org/10.21037/tcr-20-1715).
Methods
Patients
Twenty patients (mean age, 64 y, age range, 46–81 y; 12 men, 8 women) were prospectively enrolled between July 2014 and June 2015 for this pilot study.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Domain Specific Review Board of National Healthcare Group, Singapore (Ref: 2014/00459) and informed consent was taken from all the patients. All these patients were presented with biopsy-proven primary lung cancer or suspicious abnormalities in the lung planned for definitive lung surgery. The data have been previously reported (24), focusing on investigating small lesion detectability for lung cancer screening with low FDG injections. The effect of lowering dose on RF derived from large lesions was explored in this manuscript.
Data acquisition
All subjects underwent whole-body 18F-FDG PET/CT scan on a Siemens Biograph mCT scanner with a 64-slice spiral CT (Siemens Healthcare Molecular Imaging) located in the Clinical Imaging Research Center after an uptake period of around 60 min with injection of 5.9±0.14 mCi 18F-FDG linearly with body weight. PET raw data were obtained in list-mode for 10 minutes with 2 bed positions covering the lungs. Each CT was performed with a tube voltage of 120 kV and a tube current of 50 mAs (rotation time: 0.5 s; pitch: 0.8).
Image reconstruction
The reduced PET doses were simulated by randomly discarding events in the list mode stream based on nine predefined true counts (24): 0.25×106, 0.5×106, 1×106, 2×106, 5×106, 7.5×106, 10×106, 15×106, and 20×106. The true PET counts were produced by subtracting prompts from randoms. The maximum possible number of independent realizations was determined based on the acquired counts for each subject, up to 50 realizations. Each realization was reconstructed with ordinary Poisson ordered subsets expectation maximization (OP-OSEM) (28) using a system PSF incorporated in the projection matrix and TOF information. Corrections including attenuation, randoms, and scatter were carried out for each realization. PET images were reconstructed with 2 iterations and 21 subsets and then smoothed by a 5 mm full-width-at-half-maximum Gaussian filter. The image matrix is 400×400 with a voxel size of 2.04×2.04×2.03 mm3. The dimension of CT image was 512×512 with a voxel size of 1.52×1.52×5 mm3. Low-dose PET simulations and image reconstructions were both performed on the Siemens PET/CT workstation.
Volume of interest (VOI) delineation
The VOI in the normal lung background and liver background for each subject was obtained by drawing a sphere with a diameter of 3 cm. VOIs were obtained semi-automatically by using an adaptive isocontour method correcting for local background with a threshold of 50% of the SUVpeak (29,30), following by a morphological closing operation to include necrotic regions. In addition, a manual adjustment to exclude neighboring nodes or metastases was made for each VOI if necessary. As in (22), only lesions with voxel number greater than 64 (4×4×4) were included for the meaningful calculations of RF. In addition, to mitigate the impact of partial volume effect on texture calculations, lesions with volumes with dimensions of at least equal to 3 times the spatial resolution of the scanner (about 5 mm) were included. Therefore, only the lesion with a volume of greater than 1.5 cm3 was included in this study. All of the VOIs were drawn on the PET images reconstructed with full count data and then copied to the images at reduced counts.
Image features
A bin size of 64 was selected in this study to calculate TFs (22,31). Image intensities within each VOI were transformed to the range [0–63]. For each VOI, 67 RFs were computed for each VOI from FOS, GLCM, GLRLM, GLSZM, NGLDM and NGTDM (19) and their acronyms are summarized in Table 1, including seven SUV measures [lesion SUVmax, normal lung SUVmax, normal liver SUVmax, lesion SUVmean, normal lung SUVmean, normal liver SUVmean. Lesion SUVpeak (1-cm-diameter spherical VOI centered on SUVmax)]. In terms of GCLM, 3D matrix was employed in this study, capturing spatial dependence of gray-level values across multiple slices, which was extended from the 2D GLCM by summing voxel triplet probabilities in a 2D image. Each TF of GLCM and GLRLM was calculated from the combined matrix from 13 different directions with 1 voxel displacement (32). The 26 nearest neighbors were employed to calculate NGLDM and NGTDM. Moreover, the neighborhood intensity difference of zero was only considered for NGTDM. All of the TFs were calculated based on an open-source software package (33) following the guideline of image biomarker standardization initiative (34). The VOI delineations and RF calculations were performed on an in-house software written with Matlab running on a normal desktop computer.
Table 1
| Order | Matrix | Features |
|---|---|---|
| FOS | Histogram | Variance (VAR) |
| Median (MED) | ||
| Coefficient of variation (COV) | ||
| Skewness (SKE) | ||
| Kurtosis (KUR) | ||
| Energy (ENEF) | ||
| Entropy (ENTF) | ||
| Lesion_mean (LSM) | ||
| Lung_mean (LUM) | ||
| Liver_mean (LVM) | ||
| Lesion_max (LSX) | ||
| Lung_max (LUX) | ||
| Liver_max (LVX) | ||
| Lesion_peak (LSP) | ||
| Second-order features | GLCM | Autocorrelation (AUC) |
| Contrast (CONG) | ||
| Correlation (COR) | ||
| Cluster shade (CS) | ||
| Dissimilarity (DIS) | ||
| Energy (ENG) | ||
| Entropy (ENTG) | ||
| Inverse difference (ID) | ||
| Homogeneity (HM) | ||
| Maximum probability (MP) | ||
| Sum of squares (SOS) | ||
| Sum average (SA) | ||
| Sum variance (SV) | ||
| Sum entropy (SE) | ||
| Difference variance (DV) | ||
| Difference entropy (DE) | ||
| Information measure of correlation (IMC) | ||
| Inverse difference normalized (IDN) | ||
| Inverse difference moment normalized (IDMN) | ||
| Diagonal moment (DM) | ||
| Second diagonal moment (SDN) | ||
| High-order features | GLRLM | Short-run emphasis (SRE) |
| Long-run emphasis (LRE) | ||
| Low grey-level run emphasis (LGRE) | ||
| High grey-level run emphasis (HGRE) | ||
| Short-run low grey-level emphasis (SRLGE) | ||
| Short-run high grey-level emphasis (SRHGE) | ||
| Long-run low grey-level emphasis (LRLGE) | ||
| Long-run high grey-level emphasis (LRHGE) | ||
| Grey-level non-uniformity for run (GLNr) | ||
| Run-length non-uniformity (RLN) | ||
| Run percentage (RP) | ||
| GLSZM | Short-zone emphasis (SZE) | |
| Long-zone emphasis (LZE) | ||
| Low grey-level zone emphasis (LGZE) | ||
| High grey-level zone emphasis (HGZE) | ||
| Short-zone low grey-level emphasis (SZLGE) | ||
| Short-zone high grey-level emphasis (SZHGE) | ||
| Long-zone low grey-level emphasis (LZLGE) | ||
| Long-zone high grey-level emphasis (LZHGE) |
||
| Grey-level non-uniformity for zone (GLNz) | ||
| Zone-length non-uniformity (ZLN) | ||
| Zone percentage (ZP) | ||
| NGLDM | Small number emphasis (SNE) | |
| Large number emphasis (LNE) | ||
| Number nonuniformity (NN) | ||
| Second moment (SM) | ||
| Entropy (ENTN) | ||
| NGTDM | Coarseness (COA) | |
| Contrast (CONN) | ||
| Busyness (BUSN) | ||
| Complexity (COMP) | ||
| Texture strength (TS) |
TF, texture feature; FOS, first-order statistics; GLCM, gray-level co-occurrence matrix; GLRLM, gray-level run length matrix; GLSZM, gray-level size zone matrix; NGLDM, neighboring gray-level dependence matrix; NGTDM, neighbor gray-tone difference matrix.
Data analysis
Mean images of each subject across realizations were calculated for analysis at each count level. Bias percentage (BP) at different low count levels for each RF relative to its value at full count level was calculated as follows:
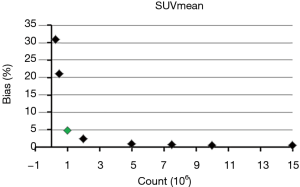
where RFlow and RFfull are the value of each RF at each low count level and full count level calculated from each mean image, respectively. The mean BP of all lesions for each RF was calculated for further analysis. Eight scales of BP were used in this study (5%, 10%, 15%, 20%, 25%, 30%, 40% and 50%). The minimum count required to reach the least BP scale for each RF was determined. For example, as for SUVmean, the mean BP were 31%, 21%, 4.8%, 2.5%, 1%, 0.7%, 0.6%, 0.5% and 0.5% at the count level of 0.25×106, 0.5×106, 1×106, 2×106, 5×106, 7.5×106, 10×106, 15×106, and 20×106, respectively, as shown in Figure 1. Therefore, the minimum count of 1×106 and the least BP scale of 5% were recorded for SUVmean.
Results
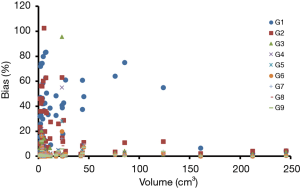
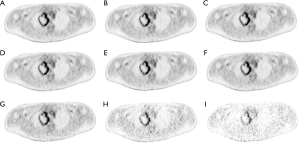
Patient characteristics were shown in Table 2. All patients were at the advanced stage (III and IV) with 16 adenocarcinoma and 4 squamous cell carcinoma. Among these patients, the number of Chinese, Malay, Filipino and Indian are 15, 3, 1 and 1, respectively. Fifty-seven lesions with a volume greater than 1.5 cm3 were identified for all 20 patients. The mean lesion volume was 25.7 (range, 1.5–245.4) cm3. Figure 2 shows PET images of a patient reconstructed with full count and different reduced count data. As expected, image quality decreases with lowering counts. The increasing noise in the image may translate into less stable image features. SUVmean bias of all lesions at different count level is displayed in Figure 3, which demonstrates that bias decreases with higher lesion volumes. The minimum count required to reach the least BP scale for each image feature is shown in Figure 4. Since the BP of all image features at the count level of 0.25×106 were larger than 50%, the information for this count level were not included in Figure 4. The SUVmean (lesion, normal lung and liver) were very stable at all count levels. Only a count of 1×106 was required to reach 5% of BP for SUVmean. In comparison with SUVmean, SUVpeak (LSP) and SUVmax (LSX, LUX and LVX) of lesion (LSX), normal lung (LUX) and liver (LVX) were less table. Counts of 5×106 and 10×106 were necessary to maintain 5% of BP for LSP and LSX, respectively. BP was found to be higher for LUX (20%) and LVX (40%) even at the count of 20×106. Textures including entropy (GLCM), sum entropy (GLCM), busyness (NGTDM) and RLN (GLRLM) were the most stable RFs, having the same performance with SUVmean. In addition, MED, KURENEF and ENTF from FOS, ZLN and ENG from GLCM, ZP from GLSZM, TS from NGTDM, GLNr, LRE and RP from GLRLM can achieve a BP of 5% at the count of not more than 5×106. A count level of 20×106 was required to achieve a BP of 5% for features including ID(GLCM), HM(GLCM), IMC(GLCM), IDN(GLCM), IDMN(GLCM), DM(GLCM), GLNz(GLSZM), SZE(GLSZM) and SRE(NGLD). CS(GLCM), LRLGE(GLRM), HGRE(GLRM) and SRLGE(GLRM) exhibited the worst performance, their BP were more than 50% even with 20×106 counts. The count of 1×106 is approximate to an effective dose of around 0.04 mSv.
Table 2
| Subject | Sex | Race | Age (y) | Histology | TNM stage |
|---|---|---|---|---|---|
| 1 | Male | Indian | 56 | Squamous cell carcinoma | IIIB |
| 2 | Female | Chinese | 47 | Squamous cell carcinoma | III |
| 3 | Male | Filipino | 52 | Adenocarcinoma | IV |
| 4 | Male | Chinese | 66 | Adenocarcinoma | IV |
| 5 | Male | Malay | 66 | Adenocarcinoma | IV |
| 6 | Female | Malay | 54 | Adenocarcinoma | IV |
| 7 | Female | Chinese | 70 | Adenocarcinoma | IV |
| 8 | Male | Chinese | 80 | Adenocarcinoma | IV |
| 9 | Female | Chinese | 59 | Adenocarcinoma | IV |
| 10 | Male | Chinese | 68 | Squamous cell carcinoma | IV |
| 11 | Female | Chinese | 56 | Adenocarcinoma | IV |
| 12 | Male | Chinese | 46 | Adenocarcinoma | IV |
| 13 | Female | Chinese | 74 | Adenocarcinoma | IV |
| 14 | Male | Chinese | 71 | Adenocarcinoma | IV |
| 15 | Female | Chinese | 80 | Adenocarcinoma | IB |
| 16 | Female | Chinese | 56 | Adenocarcinoma | IV |
| 17 | Male | Chinese | 64 | Adenocarcinoma | IV |
| 18 | Male | Chinese | 81 | Adenocarcinoma | III |
| 19 | Male | Chinese | 58 | Squamous cell carcinoma | IIIB |
| 20 | Male | Malay | 68 | Adenocarcinoma | IV |

Discussion
In the last years, RF, mostly TF, have been growingly used in the quantification of intra-tumor heterogeneity. The utility of 18F-FDG PET based radiomics have been investigated in cancer patients, mainly focusing on diagnosis, treatment response prediction and prognosis. The workflow of radomics typically involves quantitative imaging, VOI delineation, RF extraction and machine learning, in which error from each step would lead to variations of radiomics performance. PET imaging is an intrinsically noisy imaging modality and its quantitative image reconstruction easily suffers from factors including physical, technical and biological factors (14). Detected PET count or events during acquisition usually depends on injected tracer dose, system sensitivity and scan duration, which determines PET image quality and has influences on accuracy of RF. Our primary aim was to explore the impact of reducing count on RFs derived from 18F-FDG PET tumor images. In particular, we want to find out which RF is not sensitive to noise due to lowering PET true count level. RF with the same robustness as the commonly used SUV measures are desired for quantitative tumor characterization in low-dose PET/CT imaging, which may bring more benefit for pediatric patients or patients who need to have multiple PET/CT scans.
In a previous study (24), quantitative accuracy and good lesion detectability of low-dose 18F-FDG PET focusing on small lesions (mean volume, 1.25 cm3; range, 0.18–3.80 cm3) could be maintained around 10×106 true counts using a dataset of biopsy-proven primary lung cancer or patients with suggestive radiologic abnormalities planned for definitive lung surgery from a prospective study. Low-dose data were produced by randomly discarding events in the PET list-mode. The same image data were used in this study but focusing on large lesions (mean volume, 25.7 cm3, volume range, 1.5–245.4 cm3). Previous studies investigated the minimal size required for radiomic analysis to assess intratumor heterogeneity (32,35,36). For example, Hatt et al. suggested a size of at least 10 cm3 to evaluate tracer uptake variation with TFs (32). In that study, PET images were reconstructed with 4×4×4 mm. Therefore, number of voxels within a lesion of 1.5 cm3 in this study (voxel size: 2.04×2.04×2.03 mm) is comparable to that study. As shown in Figure 4, obviously, different image features have different sensitivity to count level. SUVmean is a very stable image metric and its BP is about 5% at the count level of 1×106, which translates to an effective dose of around 0.04 mSv. Among RF, ENTG(GLCM), SE(GLCM), BUSN(NGTDM) and RLN(GLRLM) have the same performance with SUVmean. As expected, the performance of SUVpeak(LSP) is inferior to SUVmean(LSM) but superior to SUVmax(LSX). At the count level of 20×106, BP could be to 5% for the image features including SUVmean in the lesions, normal lungs and livers, ENTG(GLCM), SE(GLCM), BUSN(NGTDM), RLN(GLRLM), MED(FOS), ENG(GLCM), ZP(GLSZM), LSP, KUR(FOS), ENEF(FOS), TS(NGTDM), GLNr(GLRLM), LRE(GLRM), RP(GLRLM), ENTF(FOS), MP(GLCM), DE(GLCM), ENTN(NGLDM), SNE(NGLDM), SUVmax in the lesions (LSX), NN(NGLDM), SM(NGLDM), LZE(GLSZM), ID(GLCM), HM(GLCM), IMC(GLCM), IDN(GLCM), IDMN(GLCM), DM(GLCM), SZE(GLSZM) and SRE(GLRLM). The effective dose is around 0.8 mSv for 20×106 counts, which is much less than that in most clinical scans (around 6 mSv). Image features including CS(GLCM), LRLGE(GLRM), HGRE(GLRM) and SRLGE(GLRM) exhibited the worst performance with BP not less than 50% with 20×106 counts. In this study, RF exhibited different variability to lowering count. However, it is not known whether the variability will be translated into significant impact in clinical practice, which will be an interesting investigation.
Discretization is usually required to reduce intensity values to a finite set and image noise for the calculation of RF. Orlhac et al. (22) suggested at least 32 gray levels should be used in the computation of RF. Our previous study (19) found that there was no significant difference for the results with a bin size of 32, 64 and 128. In addition, a fixed bin size of 0.5 SUV were also investigated in this study as recommended in the previous studies (31,37), but without changing sensitivities of image features to noise. However, choosing the optimal bin size is challenging, which depends on the noise and resolution of the PET images. Different bin size may be considered for different aims. In order to achieve the balance between accuracy and precision, the bin size of 64 was used to calculate RF.
Lesion segmentation is a prerequisite for calculations of RF. However, accurate lesion segmentation in PET is still very challenging, especially for low signal-to-noise ratio image. In this study, an adaptive contouring method with a threshold of 50% SUVpeak while correcting for local background was used to delineate VOI in the PET images with full count data and VOIs were copied to the PET images at the reduced count level to eliminate the difference due to lesion segmentation. In addition, robustness of ten conventional features including SUV measures (LSM, LUM, LVM, LSX, LUX, LVX and LSP), metabolic tumor volume, total lesion glycolysis and asphericity to noise were also investigated, while using the adaptive threshold of 50% SUVpeak on each individual image for all count levels. LSX, LUX and LVX exhibited greater than 50% variation even at the count level of 20×106. LSM, LUM, LVM, tumor volume, aspericity (38) and total lesion glycolysis were the most robust features, requiring 15×106 count to reach the threshold of 15%. A robust and accurate tumor segmentation method for low-dose PET imaging is desired.
PSF can improve isotropic PET resolution by correcting parallax effect. Although PET reconstruction with PSF modeling increases uptake measurement in small lesions and lesion detectability, it may potentially lead to distinct noise texture and cause edge overshooting (39,40). In the low-dose PET imaging, image noise can easily be blown up due to PSF modeling, which may cause instability of RF when lowering injected dose. Our previous work also demonstrated that there are quantitative differences of RF between PET images reconstruction with PSF and without PSF (19). Another major factor influencing PET radiomics for lung cancer patients is respiratory motion, since it will lead to image blurring and eventually inaccurate quantification (18). However, this motion will systematically influence each patient’s images at the all count levels and lead to the same impact on RF for each patient. Therefore, respiratory motion is not an issue for our objective in this study.
Conclusions
In this study, we analyzed the sensitivity of RF and SUV measures to count level. Different RF had different performance. The BP of SUVmean could be maintained at around 5% even at the count level of 1×106 with the patient data and acquisition parameters used in this work. ENTG(GLCM), SE(GLCM), BUSN(NGTDM) and RLN(GLRLM) have the same performance with SUVmean. MED, KURENEF and ENTF from FOS, ZLN and ENG from GLCM, ZP from GLSZM, TS from NGTDM, GLNr, LRE and RP from GLRLM can achieve a BP of 5% at the count of not more than 5×106. These features are suggested for 18F-FDG PET based radiomics study for lung cancer at true count as low as 5×106.
Acknowledgments
Funding: This study has received funding by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1715
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1715
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1715). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Domain Specific Review Board of National Healthcare Group, Singapore (Ref: 2014/00459) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Limkin EJ, Sun R, Dercle L, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol 2017;28:1191-206. [Crossref] [PubMed]
- Tang X. Texture information in run-length matrices. IEEE Trans Image Process 1998;7:1602-9. [Crossref] [PubMed]
- Thibault G, Fertil B, Navarro C, et al. Texture indexes and gray level size zone matrix. Application to cell nuclei classification. In: 10th International Conference on Pattern Recognition and Information Processing, 2009:140-5.
- Sun C, Wee GW. Neighboring gray level dependence matrix for texture classification. Comput Vis Graph Image Process 1983;23:341-52. [Crossref]
- Amadasum M, King R. Texture features corresponding to textural properties. IEEE Trans Syst Man Cybern 1989;19:1264-74. [Crossref]
- Lodge MA, Chaudhry MA, Wahl RL. Noise considerations for PET quantification using maximum and peak standardized uptake value. J Nucl Med 2012;53:1041-7. [Crossref] [PubMed]
- Hyun SH, Kim HS, Choi SH, et al. Intratumoral heterogeneity of (18)F-FDG uptake predicts survival in patients with pancreatic ductal adenocarcinoma. Eur J Nucl Med Mol Imaging 2016;43:1461-8. [Crossref] [PubMed]
- Pyka T, Gempt J, Hiob D, et al. Textural analysis of pre-therapeutic [18F]-FET-PET and its correlation with tumor grade and patient survival in high-grade gliomas. Eur J Nucl Med Mol Imaging 2016;43:133-141. [Crossref] [PubMed]
- Ohri N, Duan F, Snyder BS, et al. Pretreatment 18F-FDG PET textural features in locally advanced non-small cell lung cancer: secondary analysis of ACRIN 6668/RTOG 0235. J Nucl Med 2016;57:842-8. [Crossref] [PubMed]
- Beukinga RJ, Hulshoff JB, van Dijk LV, et al. Predicting response to neoadjuvant chemoradiotherapy in esophageal cancer with textural features derived from pretreatment 18F-FDG PET/CT imaging. J Nucl Med 2017;58:723-9. [Crossref] [PubMed]
- Cheng NM, Fang YH, Chang JT, et al. Textural features of pretreatment 18F-FDG PET/CT images: prognostic significance in patients with advanced T-stage oropharyngeal squamous cell carcinoma. J Nucl Med 2013;54:1703-9. [Crossref] [PubMed]
- Sollini M, Cozzi L, Antunovic L, et al. PET radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Sci Rep 2017;7:358. [Crossref] [PubMed]
- Hatt M, Tixier F, Pierce L, et al. Characterization of PET/CT images using texture analysis: the past, the present... any future? Eur J Nucl Med Mol Imaging 2017;44:151-65. [Crossref] [PubMed]
- Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nucl Med 2009;50:11S-20S. [Crossref] [PubMed]
- Boellaard R. Need for standardization of 18F-FDG PET/CT for treatment response assessments. J Nucl Med 2011;52:93S-100S. [Crossref] [PubMed]
- Yip S, McCall K, Aristophanous M, et al. Comparison of texture features derived from static and respiratory-gated PET images in non-small cell lung cancer. PLoS One 2014;9:e115510. [Crossref] [PubMed]
- Oliver JA, Budzevich M, Zhang GG, et al. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol 2015;8:524-34. [Crossref] [PubMed]
- Grootjans W, Tixier F, van der Vos CS, et al. The impact of optimal respiratory gating and image noise on evaluation of intratumor heterogeneity on 18F-FDG PET imaging of lung cancer. J Nucl Med 2016;57:1692-8. [Crossref] [PubMed]
- Yan J, Chu-Shern JL, Loi HY, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med 2015;56:1667-73. [Crossref] [PubMed]
- Galavis PE, Hollensen C, Jallow N, et al. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010;49:1012-6. [Crossref] [PubMed]
- Hatt M, Tixier F, Cheze Le Rest C, et al. Robustness of intratumour 18F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging 2013;40:1662-71. [Crossref] [PubMed]
- Orlhac F, Soussan M, Maisonobe JA, et al. Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med 2014;55:414-22. [Crossref] [PubMed]
- Siegel JA, Pennington CW, Sacks B. Subjecting radiologic imaging to the linear no-threshold hypothesis: a non sequitur of non-trivial proportion. J Nucl Med 2017;58:1-6. [Crossref] [PubMed]
- Schaefferkoetter JD, Yan J, Sjoholm T, et al. Quantitative accuracy and lesion detectability of low-dose 18F-FDG PET for lung cancer screening. J Nucl Med 2017;58:399-405. [Crossref] [PubMed]
- Schaefferkoetter JD, Yan J, Townsend DW, et al. Initial assessment of image quality for low-dose PET: evaluation of lesion detectability. Phys Med Biol 2015;60:5543-56. [Crossref] [PubMed]
- Yan J, Schaefferkoette J, Conti M, et al. A method to assess image quality for low-dose PET: analysis of SNR, CNR, bias and image noise. Cancer Imaging 2016;16:26. [Crossref] [PubMed]
- von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med 2007;4:e296. [Crossref] [PubMed]
- Panin VY, Kehren F, Michel C, et al. Fully 3-D PET reconstruction with system matrix derived from point source measurements. IEEE Trans Med Imaging 2006;25:907-21. [Crossref] [PubMed]
- Frings V, van Velden FH, Velasquez LM, et al. Repeatability of metabolically active tumor volume measurements with FDG PET/CT in advanced gastrointestinal malignancies: a multicenter study. Radiology 2014;273:539-48. [Crossref] [PubMed]
- van Velden FH, Kramer GM, Frings V, et al. Repeatability of radiomic features in non-small-cell lung cancer [(18)F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol 2016;18:788-95. [Crossref] [PubMed]
- Leijenaar RT, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015;5:11075. [Crossref] [PubMed]
- Hatt M, Majdoub M, Vallieres M, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med 2015;56:38-44. [Crossref] [PubMed]
- Fang YH, Lin CY, Shih MJ, et al. Development and evaluation of an open-source software package "CGITA" for quantifying tumor heterogeneity with molecular images. Biomed Res Int 2014;2014:248505.
- Alex Zwanenburg SL, Martin Vallières, Steffen Löck. Image biomarker standardisation initiative. arXiv preprint 2016;arXiv:1612.07003.
- Brooks FJ, Grigsby PW. The effect of small tumor volumes on studies of intratumoral heterogeneity of tracer uptake. J Nucl Med 2014;55:37-42. [Crossref] [PubMed]
- Forgacs A, Pall Jonsson H, Dahlbom M, et al. A study on the basic criteria for selecting heterogeneity parameters of F18-FDG PET images. PLoS One 2016;11:e0164113. [Crossref] [PubMed]
- Foley KG, Hills RK, Berthon B, et al. Development and validation of a prognostic model incorporating texture analysis derived from standardised segmentation of PET in patients with oesophageal cancer. Eur Radiol 2018;28:428-36. [Crossref] [PubMed]
- Apostolova I, Ego K, Steffen IG, et al. The asphericity of the metabolic tumour volume in NSCLC: correlation with histopathology and molecular markers. Eur J Nucl Med Mol Imaging 2016;43:2360-73. [Crossref] [PubMed]
- Tong S, Alessio AM, Kinahan PE. Noise and signal properties in PSF-based fully 3D PET image reconstruction: an experimental evaluation. Phys Med Biol 2010;55:1453-73. [Crossref] [PubMed]
- Munk OL, Tolbod LP, Hansen SB, et al. Point-spread function reconstructed PET images of sub-centimeter lesions are not quantitative. EJNMMI Phys 2017;4:5. [Crossref] [PubMed]