Application of next-generation sequencing in resistance genes of neoadjuvant chemotherapy for esophageal cancer
Introduction
Esophageal cancer (EC) is the sixth leading cause of cancer-related death worldwide (509,000 deaths in 2018), mainly categorized into adenocarcinoma (AC) and squamous cell carcinoma (SCC) (1), which differ in their pathology, tumor location, and prognosis. In contrast to other tumors, the esophagus has rich blood supply and regional lymph node drainage and is closely related to the adjacent anatomy, has a proclivity for earlier lymphatic spread and is associated with a poorer prognosis (2). Patients with EC who underwent surgery as the sole treatment had a median survival ranging from 13 to 19 months and a 5-year survival rate from 15% to 24% (3). The complex features of EC lead to the loss of the best surgical treatment opportunity, and its high invasiveness leads to a generally shorter survival period for EC patients.
Treatment of EC remains challenging but is best approached by a multidisciplinary team (4). With the use of neoadjuvant chemotherapy (NAC) and neoadjuvant chemoradiotherapy (nCRT) in the field of EC, more than half of the patients who received platinum-based neoadjuvant therapy have clinical benefit, which is significantly improved compared with the benefits seen from surgery alone (5-7). However, nearly 40% of EC patients were still not sensitive to chemotherapy, and some patients experienced disease progression during treatment (8). The underpinning mechanism of such resistance remains obscure.
Recently, advances have been achieved in the sequencing of tumor DNA (9,10). Moreover, mutations related to drug resistance were also gradually detected by the sequencing of patient samples with diseases such as lung cancer (11), breast cancer (12) and cytomegalovirus (13). Genes related to chemotherapy resistance have been discovered by the drug-resistant EC cell lines in vitro culture (14,15). However, due to the limitations of in vitro experiments, it is difficult to accurately determine the key genes mediating chemotherapy resistance. In addition, there are few reports on genome sequencing of surgical samples after NAC to study chemotherapy resistance in EC patients.
To this end, we conducted a cohort of 13 NAC patients with different stages and chemotherapy responses to monitor the gene mutations of these patients by using next-generation sequencing (NGS) and screened some high-frequency mutant genes for further analysis to reveal possible resistance genes in patients with EC. We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-322).
Methods
Patients
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Participants gave written informed consent before the study, and the study protocol was approved by the Ethics Review Committee of Henan Cancer Hospital/The Affiliated Cancer Hospital of Zhengzhou University with approval number 2014ys38. Patient details were collected on the first day of their hospitalization, as were pretreatment examinations, treatment and follow-up data. The inclusion criteria of this study were as follows: consecutive patients with thoracic EC from November 2014 to January 2015; age 20 to 75 years old; thoracic EC; pathological T stage 1–3 according to the TNM classification of malignant tumors 8th edition (16); laboratory tests confirming that bone marrow, liver and kidney function can tolerate chemotherapy; no history of other malignant tumors; surgical treatment can achieve radical resection; and Eastern Cooperative Oncology Group (ECOG) physical status score of 0 to 2. Each patient completed the following preoperational tests: electronic ultrasound gastroscopy with pathological examination, enhanced thoracic and upper abdominal computed tomography (CT) scan, upper gastrointestinal contrast imaging, abdominal and cervical color ultrasound, emission CT (ECT), pulmonary function test, electrocardiography and other routine tests. If positron emission tomography-CT (PET/CT) was accepted by patients, it was done.
Treatment
NAC comprised two cycles. Cisplatin or nedaplatin plus paclitaxel were repeated once every 3 weeks. A dose of 175 mg/m2 paclitaxel was given by intravenous drip infusion on day 1. Cisplatin or nedaplatin was administered at a dose of 75 mg/m2 by continuous infusion on days 2 to 4. Two to 6 weeks after NAC, right a thoracolaparoscopic esophagectomy and at least 2-field lymph node dissections were performed. CT was used to evaluate the effects of all patients after every therapy cycle. The Response Evaluation Criteria in the Solid Tumors (RECIST) were used to evaluate the clinical tumor responses (17). According to the international RECIST criteria, the thoracic CT was evaluated after each-cycle chemotherapy as follows: partial response (PR): the total length of all baseline target lesions decreased by ≥30%; stable disease (SD): all baseline target lesions did not reach PR or increased to progress disease (PD); PD: the total length of all baseline target lesions increased by ≥20%, or one or more new lesions appeared; or complete response (CR): all lesions disappeared, no thickening of the esophageal wall <5 mm (18), the original enlarged lymph nodes disappeared, no new lesions.
DNA extraction
Tumor tissue samples were obtained from postoperative formalin-fixed and paraffin-embedded (FFPE) tissue of 13 patients. Fifteen mg of tumor tissue was isolated from the obvious tumor infiltrated area of each sample. Then genomic DNA was extracted with the QIAamp DNA Mini Kit (Qiagen, Germany) by repeated centrifugation and purification according to the manufacturer’s instructions. And the same DNA extraction process was performed on the prechemotherapy gastroscope FFPE tumor samples of four patients and mononuclear cell from 3 mL blood samples of 13 patients.
Experimental procedures
Mononuclear cell DNA extracted from 3 mL blood samples of 13 patients were used to match tumor samples and control DNA quality by using a high sensitivity DNA bioanalyzer. Genetic profiles of all tissue samples were assessed by performing capture-based targeted deep sequencing using the OncoScreen panel (Burning Rock Biotech Ltd., Guangzhou, China), covering 2.02 MB of human genomic regions, including all exons and critical introns of 295 genes. The mutations related to drug resistance were screened after the consistency evaluation with the chemotherapy efficacy. Finally, the spatial conformation was performed by using Coot software (Crystallographic Object-Oriented Toolkit, Coot) (0.8.7/November 8, 2016) and PyMOL software to further screen the drug resistance genes.
Results
Patient information and effects after NAC
To investigate potential gene mutations associated with resistance in patients with EC, we used a cohort of 13 EC patients who received NAC with different chemotherapy responses and sequenced the biopsy specimens to determine resistance genes of EC (Table 1). In the study cohort, 12 patients completed two cycles of NAC and underwent surgical resection within the next 2 weeks. Another patient with dual-source cancer received surgical treatment immediately after the first cycle of chemotherapy because of grade 3 chemotherapy-related adverse reactions. Of the 13 patients, the median age was 60.6 years, and 11 patients had a history of smoking. In the 13 patients who received NAC with platinum-based chemotherapy, 2 patients achieved a CR (15.4%), 6 patients achieved a PR (46.1%), and 5 patients had SD (38.5%).
Table 1
| Patient number | Age | Sex | Tumor location | Pathological type | Clinical stage | Pathologic stage | Stage | Histological grade | Response | OS (months) |
|---|---|---|---|---|---|---|---|---|---|---|
| N01 | 50 | Male | Upper | ESCC | cT3N1M0 | yPT1N0M0 | I | 3 | CR | 54.8 |
| N02 | 53 | Male | Middle | ESCC | cT3N0M0 | yPT1N0M0 | I | 2 | CR | 54.0 |
| N03 | 45 | Male | Middle | ESCC | cT2N0M0 | yPT2N1M0 | IIIA | 3 | PR | 57.0 |
| N04 | 65 | Male | Middle | ESCC | cT2N0M0 | yPT2N0M0 | I | 3 | PR | 53.4 |
| N05 | 65 | Female | Middle | ESCC | cT2N0M0 | yPT2N1M0 | IIIA | 3 | PR | 57.5 |
| N06 | 68 | Male | Lower | EAC | cT3N1M0 | yPT3N2M0 | IIIB | 3 | PR | 60.0 |
| N07 | 65 | Male | Middle | ESCC | cT2N0M0 | yPT2N0M0 | I | 3 | PR | 59.0 |
| N08 | 60 | Male | Middle | ESCC | cT2N0M0 | yPT2N0M0 | I | 2 | PR | 57.8 |
| N09 | 66 | Male | Middle | ESCC | cT3N0M0 | yPT3N0M0 | II | 2 | SD | 57.3 |
| N10 | 59 | Male | Upper and middle | ESCC | cT3N2M0 | yPT2N3M0 | IVA | 3 | SD | 7.9 |
| N11 | 70 | Female | Middle | ESCC | cT2N0M0 | yPT3N0M0 | II | 3 | SD | 55.3 |
| N12 | 59 | Female | Middle | ESCC | cT2N1M0 | yPT2N0M0 | I | 3 | SD | 57.3 |
| N13 | 63 | Male | Middle | ESCC | cT2N0M0 | yPT2N0M0 | I | 3 | SD | 46 |
ESCC, esophageal squamous cell carcinoma; EAC, esophageal adenocarcinoma; CR, complete response; PR, partial response; SD, stable disease; OS, overall survival.
Statistics and results of NGS in tissues after chemotherapy
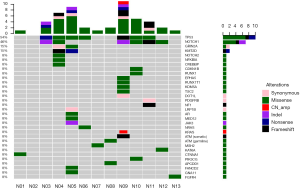
Among the 13 postoperative samples, 11 tissue samples were successfully paired with the blood cell samples, and sequencing was completed. Two samples failed, and the pairing was successful after replenishing the tissue samples (N06, N13). In our NGS results (Figure 1), the median sequencing depth of all samples was >500, which was consistent with the expected sequencing depth. All sample quality tests were qualified before sequencing, and a total of 36 somatic mutations were detected in 8 cases, including missense (21 cases, 58.3%), frameshift (5 cases, 13.9%), synonymous (4 cases, 11.1%), and nonsense mutations (2 cases, 5.6%), indels (3 cases, 8.3%) and copy number amplification (1 case, 2.8%). A total of nine likely pathogenic germline mutations were detected, all of which were missense mutations.
Based on the NGS results, 7 (54%) samples harbored somatic mutations of TP53, and 6 samples (46%) harbored somatic mutations of NOTCH1. In the SD group, 4/5 patients harbored the NOTCH1 mutation, 3/5 patients harbored the TP53 mutation, and the proportion in the PR group was 2/6 and 4/6. No somatic mutations were detected in two patients with CR.
Features and description of high-frequency gene mutations
The mean tumor mutational burden (TBM) of 13 samples was 4.13/Mb, which was moderate among multiple cancers (12). The mean value of the sample TBM in six patients of the PR group was 4.75/Mb, and the value in the SD group was 5/Mb. Based on whether the results showed a clinical response, we found no significant difference in the sample mutation load between the two groups [(CR + PR):SD, P=0.598]. The mutation signature in all 13 samples included C>A, C>T, C>G, A>G, A>C and A>T, and a high frequency of C>T(40%) transitions was identified overall in the patients, consistent with previously published data on esophageal SCC (ESCC) and esophageal AC (EAC) (19,20).
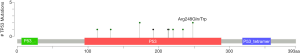
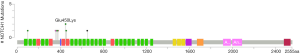
Among the 36 somatic mutations, we found that TP53 and NOTCH1 mutation were significantly elevated in the resistant patients (Table 2). A total of 11 types TP53 mutations were detected in samples from seven patients, and these mutations were mainly located in the DNA-binding domain which were closely related to carcinogenesis (Figure 2). Similarly, nine types of NOTCH1 mutations were detected, which were mainly distributed in the front-end EGF-like region of the NOTCH1 gene and concentrated in the ligand binding region (Figure 3). One patient developed three mutations gene with allele frequency (AF) over 60%; this was associated with only one cycle of NAC and the pathological features of dual-source cancer. Two patients in the PR group were found to have Glu450Lys somatic mutations, which were located in the NOTCH1 calcium-binding EGF domain, and this mutation was the only repeatedly detected NOTCH1 mutation, which was considered to be closely related to EC resistance.
Table 2
| Patient number | Gene | Mutation type | AF, % | Mutation site |
|---|---|---|---|---|
| N03 | TP53 | Missense | 8.00 | TP53:c.338T>G(p.Phe113Cys) |
| TP53 | Stop_gained | 4.30 | TP53:c.637C>T(p.Arg213*) | |
| TP53 | Missense | 3.50 | TP53:c.659A>G(p.Tyr220Cys) | |
| NOTCH1 | Missense | 8.10 | NOTCH1:c.293G>T(p.Cys98Phe) | |
| NOTCH1 | Missense | 2.70 | NOTCH1:c.1348G>A(p.Glu450Lys) | |
| NOTCH1 | Inframe_ deletion | 2.00 | NOTCH1:c.1138_1140delCCC(p.Pro380del) | |
| N04 | TP53 | Missense | 24.50 | TP53:c.518T>G(p.Val173Gly) |
| NOTCH1 | Missense | 2.90 | NOTCH1:c.1120G>T(p.Asp374Tyr) | |
| NOTCH1 | Missense | 3.10 | NOTCH1:c.1348G>A(p.Glu450Lys) | |
| N05 | TP53 | Missense | 11.80 | TP53:c.701A>G(p.Tyr234Cys) |
| N06 | TP53 | Missense | 2.70 | TP53:c.641A>G(p.His214Arg) |
| N09 | TP53 | Stop_ gained | 2.20 | TP53:c.574C>T(p.Gln192*) |
| TP53 | Missense | 1.10 | TP53:c.742C>T(p.Arg248Trp) | |
| TP53 | Frameshift_variant | 18.60 | TP53:c.516dupT(p.Val173fs) | |
| NOTCH1 | Disruptive Inframe_ deletion | 26.40 | NOTCH1:c.1070_1072delTCT(p.Phe357del) | |
| N10 | TP53 | Missense | 63.70 | TP53:c.743G>A(p.Arg248Gln) |
| NOTCH1 | Missense | 78.50 | NOTCH1:c.1366T>A(p.Cys456Ser) | |
| NOTCH1 | Missense | 77.20 | NOTCH1:c.1365G>T(p.Glu455Asp) | |
| N11 | TP53 | Missense | 6.00 | TP53:c.396G>T(p.Lys132Asn) |
| NOTCH1 | Frameshift_variant | 8.00 | NOTCH1:c.1952_1958delACTCGGG(p.Asp651fs) | |
| N12 | NOTCH1 | Missense | 2.40 | NOTCH1:c.1093C>T(p.Arg365Cys) |
Frameshift_variant: insertion or deletion causes a frame shift. Missense: variant causes a codon that produces a different amino acid. Stop_gained: variant causes a STOP codon (nonsense_variant). Inframe_deletion: one or many codons are deleted. Disruptive inframe_deletion: one codon is changed and one or more codons are deleted. AF, allele frequency.
Comparison of gene mutations before and after chemotherapy
According to different curative effects, we tracked the prechemotherapy tumor tissue of four patients and performed NGS on these four patients, including two patients with SD, one with PR, and one with CR (Figure 4). By comparing somatic gene mutations in the four patients before and after chemotherapy, we found that all of the somatic mutations detected in patients N02 (Figure 4A) before chemotherapy disappeared, and the clinical effect was CR, and there was no new gene mutation after two cycles of NAC.
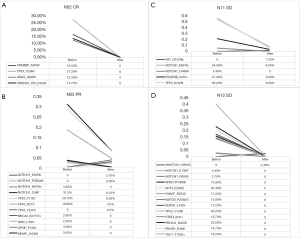
Comparing the NGS results before and after chemotherapy in patient N03 (Figure 4B), the AF of NOTCH1_C98F, TP53_F113C and TP53_R213*, which was detected before NAC, decreased significantly after chemotherapy, and the residual mutation disappeared after chemotherapy, while NOTCH1_E450K, NOTCH1_P380del and TP53_y220C were the newly identified mutations during chemotherapy. Similarly, the high AF of NOTCH1_D651fs, TP53_K132N and PDGFRB_S470 in patient N11 (Figure 4C) were significantly decreased after NAC, and NF1_Q1520fs was a new somatic mutation found after chemotherapy. In the sample of patient N12 (Figure 4D), we found that all the gene mutations in the tumor cells disappeared after chemotherapy, and NOTCH1_R365C was a newly detected mutation during chemotherapy. A total of five newly generated mutations were detected during NAC, and some somatic mutations did not completely disappear after two cycles of NAC.
Protein conformation analysis
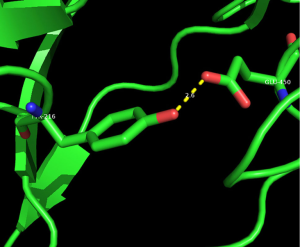
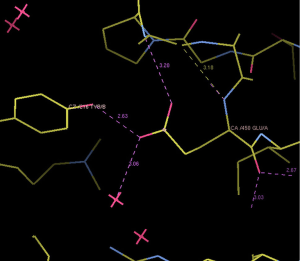
We selected three missense mutations in the vicinity of the NOTCH1 gene for protein conformational analysis, including NOTCH1:c.1093C>T (p. Arg365Cys), NOTCH1:c.1120G>T (p. Asp374Tyr), NOTCH1:c.1348G>A (p. Glu450Lys). According to the results of spatial conformation and drug resistance analysis of these three missense mutations, we excluded the unstructured coverage of Arg365Cys and Asp374Tyr. In the conformational analysis of Glu450Lys (Figure 5), we found that glutamate formed by normal transcription at this point can form a hydrogen bond with tyrosine on NOTCH1 ligand protein Delta-like 4 (DLL4) (Figure 6). Then, the newly mutated lysine may form a salt bridge structure with tyrosine after the mutation of the base sequence, resulting in a tighter connection between NOTCH1 and the ligand, making the pathway more susceptible to activation or prolonging activation time; moreover, this mutation was the only repeated missense mutation in all of the samples we studied.
Discussion
In current research, most of the studies on chemoresistance related to EC focus on the expression of cell surface receptors and ligands, and the report of genome sequencing of EC was also focused mainly on the relationship between gene mutations and the carcinogenesis or prognosis of EC. The connection between gene mutations and chemotherapy resistance has not been addressed by existing research. In our study, we performed NGS analysis on 13 EC patients for the detection of NAC drug resistance mutations. Somatic mutations (9/11) were commonly found in the patients with chemotherapy response to PR and SD, and mutation genes at the same site were found in tissue samples of two patients with PR. Among the six patients with mutations in the NOTCH1 gene, four patients responded to chemotherapy with SD and two patients with PR, suggesting that the mutations in NOTCH1 gene are likely to cause chemotherapy to be relatively insensitive or ineffective to these mutant cells. In addition, most of the NOTCH1 gene mutation sites found in this study are located in the front-end EGF-like region of the NOTCH1 gene and concentrated in the ligand binding region. Mutations at this site may result in a change in the conformation of its encoded NOTCH1 homologous protein, which may lead to abnormal binding to the ligand DLL4. Thus, the NOTCH1 signaling pathway may be continuously activated, making tumor cells relatively insensitive to chemotherapy.
According to the existing research, the NOTCH signaling pathway is generally considered an evolutionarily conserved intercellular interaction mechanism, which profoundly affects cell development, proliferation, differentiation, survival and apoptotic events (21). NOTCH signals control cellular fates and the segregation of lineages by linking the fate of one cell to that of a neighbor through the interaction of the Notch surface receptor expressed on one cell with the membrane-bound ligands expressed on the surface of an adjacent cell (22). The activated NOTCH1 signals result in the release of the Notch1 intracellular domain into the nucleus and the activation of downstream genes, including Hes-1, Hey-1, and others (23). In addition, simultaneous activation of the Notch1 pathway can induce SKP2 expression and enhance the role of Notch signaling in promoting cell cycle progression by enhancing the proteasome-mediated degradation of p27kipl and p21cipl (24). In this way, NOTCH1 may be associated with dismal prognosis, early stage malignancy, and less regional lymph node metastasis, which exhibit a poor outcome and failure to respond to standard chemotherapy treatment (25).
In the studies of the chemosensitivity of prostate cancer (26) and breast cancer (23), the sensitivity of some patients to chemotherapy drugs can be improved by downregulating the NOTCH1 signaling pathway. In addition, the study of cisplatin resistance in head and neck SCC by Gu et al. (27) also suggested that high expression of NOTCH1 was associated with cisplatin resistance. This suggests that the signaling pathway mediated by the NOTCH1 receptor and its ligand is closely related to chemoresistance. In our study, patients with mutations in the NOTCH1 gene also tended to be relatively insensitive to chemotherapy, and these mutations are most likely a new genetic mutation acquired during treatment with chemotherapy. In addition, preliminary studies have shown that the Notch signaling pathway has good prospects as an antitumor target (28). Further research is needed to determine whether the sensitivity to chemotherapy can be improved after the application of the NOTCH1 pathway inhibitor in drug-resistant EC.
Comparison of gene mutations before and after chemotherapy in four patients revealed that most of the gene mutations disappeared after two cycles of NAC, and patients with a higher degree of AF had a significant decline after chemotherapy. Whether these genetic mutations that did not completely disappear can further improve the efficacy of chemotherapy by adjusting the frequency, concentration or manner of administration of these patients requires further study. Moreover, our results suggested that the gene mutation status of tumors could change during the process of platinum-based chemotherapy, which was also mentioned in the study of EGFR gene expression in lung cancer by Wang et al. (29). The impact of these emerging genetic mutations requires research on their specific molecular structures and more data sources for analysis. However, changes in gene mutations during chemotherapy alone do not unilaterally explain the different sensitivities of patients to chemotherapeutic drugs. As the basis of adjuvant therapy for EC, the premise that platinum drugs exert the spectral antitumor effect requires that they must enter the cell to form an adduct with DNA to limit the release of DNA (30) and prevent mitosis of cells, arresting them in G2 phase (31). Therefore, there are still other mechanisms, including drug transport, metabolism, concentration, etc., that have not yet been fully studied, which may result in the low chemosensitivity of patients.
Conclusions
We analyzed the effect of platinum-based NAC on gene mutations in patients with EC, and our NGS results indicated that the consequences of molecular detection can reflect the efficacy of chemotherapy. Furthermore, we provide a novel insight that the NOTCH1 somatic mutation may be related to the efficacy of NAC. However, due to limited research funding and less experience, we only completed the NGS of 13 patients after chemotherapy and 4 patients before chemotherapy. Then we plan to use qRT-PCR and Sanger generation sequencing technologies to independently validate the candidate drug resistance mutations identified above in other NAC tissue samples.
Acknowledgments
Thanks to the Department of Bioinformatics, Zhengzhou University for providing a graphical representation of protein conformation.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-322
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-322
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-322). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are responsible for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Participants gave written informed consent before the study, and the study protocol was approved by the Ethics Review Committee of Henan Cancer Hospital/The Affiliated Cancer Hospital of Zhengzhou University with approval number 2014ys38.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Tu CC, Hsu PK. The frontline of esophageal cancer treatment: questions to be asked and answered. Ann Transl Med 2018;6:83. [Crossref] [PubMed]
- Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003;349:2241-52. [Crossref] [PubMed]
- Krill T, Baliss M, Roark R, et al. Accuracy of endoscopic ultrasound in esophageal cancer staging. J Thorac Dis 2019;11:S1602-9. [Crossref] [PubMed]
- Sjoquist KM, Burmeister BH, Smithers BM, et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: an updated meta-analysis. Lancet Oncol 2011;12:681-92. [Crossref] [PubMed]
- Ando N, Kato H, Igaki H, et al. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907). Ann Surg Oncol 2012;19:68-74. [Crossref] [PubMed]
- Eyck BM, van der Wilk BJ, Lagarde SM, et al. Neoadjuvant chemoradiotherapy for resectable oesophageal cancer. Best Pract Res Clin Gastroenterol 2018;36-37:37-44. [Crossref] [PubMed]
- Zheng Y, Li Y, Liu X, et al. Reevaluation of neoadjuvant chemotherapy for esophageal squamous cell carcinoma: a meta-analysis of randomized controlled trials over the past 20 years. Medicine (Baltimore) 2015;94:e1102. [Crossref] [PubMed]
- Tran B, Dancey JE, Kamel-Reid S, et al. Cancer genomics: technology, discovery, and translation. J Clin Oncol 2012;30:647-60. [Crossref] [PubMed]
- Sasaki Y, Tamura M, Koyama R, et al. Genomic characterization of esophageal squamous cell carcinoma: Insights from next-generation sequencing. World J Gastroenterol 2016;22:2284-93. [Crossref] [PubMed]
- Li A, Yang JJ, Zhang XC, et al. Acquired MET Y1248H and D1246N mutations mediate resistance to MET inhibitors in non-small cell lung cancer. Clin Cancer Res 2017;23:4929-37. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Sahoo MK, Lefterova MI, Yamamoto F, et al. Detection of cytomegalovirus drug resistance mutations by next-generation sequencing. J Clin Microbiol 2013;51:3700-10. [Crossref] [PubMed]
- Tsutsui M, Kawakubo H, Hayashida T, et al. Comprehensive screening of genes resistant to an anticancer drug in esophageal squamous cell carcinoma. Int J Oncol 2015;47:867-74. [Crossref] [PubMed]
- Li B, Hong P, Zheng CC, et al. Identification of miR-29c and its target FBXO31 as a key regulatory mechanism in esophageal cancer chemoresistance: functional validation and clinical significance. Theranostics 2019;9:1599-613. [Crossref] [PubMed]
- Rice TW, Ishwaran H, Ferguson MK, et al. Cancer of the esophagus and esophagogastric junction: an Eighth edition staging primer. J Thorac Oncol 2017;12:36-42.
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Davies AR, Gossage JA, Zylstra J, et al. Tumor stage after neoadjuvant chemotherapy determines survival after surgery for adenocarcinoma of the esophagus and esophagogastric junction. J Clin Oncol 2014;32:2983-90. [Crossref] [PubMed]
- Zhang L, Zhou Y, Cheng C, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet 2015;96:597-611. [Crossref] [PubMed]
- Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 2013;45:478-86. [Crossref] [PubMed]
- Androutsellis-Theotokis A, Leker RR, Soldner F, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature 2006;442:823-6. [Crossref] [PubMed]
- Louvi A, Artavanis-Tsakonas S. Notch and disease: a growing field. Semin Cell Dev Biol 2012;23:473-80. [Crossref] [PubMed]
- Kang L, Mao J, Tao Y, et al. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci 2015;106:700-8. [Crossref] [PubMed]
- Dohda T, Maljukova A, Liu L, et al. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res 2007;313:3141-52. [Crossref] [PubMed]
- Song B, Cui H, Li Y, et al. Mutually exclusive mutations in NOTCH1 and PIK3CA associated with clinical prognosis and chemotherapy responses of esophageal squamous cell carcinoma in China. Oncotarget 2016;7:3599-613. [Crossref] [PubMed]
- Liu X, Luo X, Wu Y, et al. MicroRNA-34a Attenuates paclitaxel resistance in prostate cancer cells via direct suppression of JAG1/Notch1 axis. Cell Physiol Biochem 2018;50:261-76. [Crossref] [PubMed]
- Gu F, Ma Y, Zhang Z, et al. Expression of Stat3 and Notch1 is associated with cisplatin resistance in head and neck squamous cell carcinoma. Oncol Rep 2010;23:671-6. [PubMed]
- Xiao YF, Yong X, Tang B, et al. Notch and Wnt signaling pathway in cancer: Crucial role and potential therapeutic targets Int J Oncol 2016;48:437-49. (Review). [Crossref] [PubMed]
- Wang Y, Ma X, Wei Y, et al. Effect of platinum-based chemotherapy on EGFR gene mutation status in lung adenocarcinoma. Medicine (Baltimore) 2018;97:e9602. [Crossref] [PubMed]
- Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 2005;4:307-20. [Crossref] [PubMed]
- Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics 2009;1:280-91. [Crossref] [PubMed]