
MiRNA 3613-5p and MiRNA 3916 rescued the inhibition of cell migration in CNOT2 depleted MDA-MD-231 cells
Introduction
The CCR4-NOT complex (CNOT), which is composed of nine core subunits (CNOT1, CNOT2, CNOT3, CNOT6, CNOT6L, CNOT7, CNOT8, CNOT9/RQCD1, and CNOT10), is a large (>2 MDa) multisubunit, multifunctional regulator (1) with multiple cellular functions, including effects on mRNA stability (2), translation (3), and transcription mediated by RNA polymerases I and II. Furthermore, CNOT functions in transcription by repressing TATA-less core promoters. In yeast, CNOT regulates gene expression and controls both cell growth and glucose metabolism (4).
Human CNOT2 is important for maintaining the deadenylase activity and structural integrity of CNOT. CNOT2 depletion was shown to induce apoptosis in a caspase-dependent manner in HeLa cells (5), and was found to interact with cyclin-dependent kinase 11, which is cleaved by caspases (6). CNOT2 also plays multiple roles in autophagy, adipogenesis, angiogenesis, cell division, bone formation, cell proliferation, and senescence (5,7-10), and CNOT represses promoter activity (11). Furthermore, CNOT modulates the deadenylation of microRNA (miRNA or miR) and Piwi-interacting RNA-targeted mRNAs (12,13). Finally, CNOT interacts with Trinucleotide Repeat Containing 6 (TNRC6 or GW182) which regulates miRNA repression complexes (14,15).
miRNAs are small, noncoding RNAs that regulate gene expression at the post-transcriptional level by repressing mRNA translation or degradation (16). miRNAs have multiple biological functions, including cell growth, apoptosis, differentiation, and proliferation (17). In this study, we found that CNOT2 modulates mRNA level of Dicer, and DiGeorge Syndrome Critical Region 8 (DGCR8) activity and regulates miRNA expression in MDA-MB-231 cells. Also, has-miR-3613-5p and has-miR-3916 rescued the inhibition of migration from CNOT2 depleted MDA-MB-231 cell lines.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-19-2821).
Methods
Cell culture
Human breast cancer cells (MDA-MB-231, RRID: CVCL_0062) were obtained from the American Type Culture Collection (Bethesda, MD, USA) and cultured in RPMI1640 medium (Welgene, Daegu, South Korea) with 2 µM L-glutamine, 10% fetal bovine serum, and penicillin/streptomycin (WELGENE, Korea) under 5% CO2.
Real-time quantitative RT-PCR (RT-qPCR)
Total RNA from MDA-MB-231 cells was extracted using QIAzol (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized from total RNA using a Reverse Transcription Kit (Promega, Madison, WI, USA). A quantitative analysis was performed with SYBRgreen master mix and carried out using a LightCyclerTM instrument (Roche Applied Sciences, Indianapolis, IN, USA). The primers were as listed in Table 1.
Table 1
| Gene | Forward primers (5' to 3') | Reverse primers (5' to 3') |
|---|---|---|
| CNOT2 | GGTAACCCAACTCCATTAATAAACC | TGCTGGTTTTGTTACCATTCC |
| DGCR8 | GCAAGATGCACCCACAAAGA | TTGAGGACACGCTGCATGTAC |
| Dicer | TTAACCTTTTGGTGTTTGATGAGTGT | GGACATGATGGACAATTTTCACA |
| GAPDH | CCA CTC CTC CAC CTT TGA C | ACC CTG TTG CTG TAG CCA |
RNA interference
Control or CNOT2 small interfering RNA (siRNA #1. Cat 108569, siRNA#108571; Thermo Fisher Scientific, Waltham, MA, USA) was transfected into MDA-MB-231 cells using Lipofectamine Transfection Reagent (Invitrogen, USA) according to the manufacturer’s protocol.
Generation of CNOT2 short hairpin RNA (shRNA) stably transfected cells
To generate MDA-MB-231 stable cell line for depletion of CNOT2 shRNA (TRCN0000015129CCGGCGGGTTACTAACATTCCTCAACTCGAGTTGAGGAATGTTAGTAACCCGTTTTT), control (shControl; pLKO.1puro; SHC002, Sigma-Aldrich) were transfected into MDA-MD-231 cells using the Lipofetamine reagent (Thermo scientific, USA) according to the manufacturer’s protocol.
miRNA array
miRNA expression profiling was carried out using total RNA from MDA-MB-231 cells treated with CNOT2 siRNA as described previously (18). For miRNA profiling, Affymetrix GeneChip® miRNA 4.0 (Thermo Fisher Scientific Inc., USA) was used. Total RNA was extracted using QIAzol (Invitrogen, USA) according to the manufacturer’s protocol.
Wound healing assay
miRNA mimics (Genolution, Korea) were transfected using INTERFER in siRNA transfection reagent (Polyplus-transfection, USA). After 48 transfection, a wound was generated using a sterile plastic pipette tip and were incubated for 24 h. The cells were fixed and stained with 1% crystal violet (Sigma-Aldrich, USA). To visualize the wound area, an inverted microscope was used.
Pathway analysis
To see the function of differentially expressed miRNAs, target of miRNAs were used for the Protein Analysis Through Evolutionary Relationship (PANTHER) classification system and analysis tools.
Statistical analysis
All data are expressed as the mean ± standard error of the mean (SEM). The data were analyzed using one-way analysis of variance (one-way ANOVA) with SigmaPlot (SYSTAT Software Inc., Chicago, IL, USA).
Results
CNOT2 regulates Dicer, and DGCR8
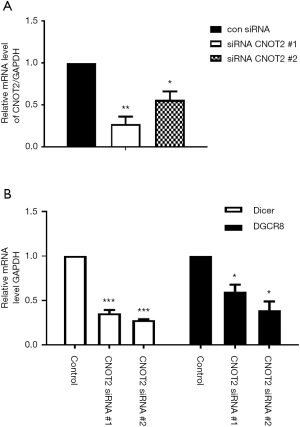
To determine whether CNOT2 affects miRNA biogenesis, we examined the mRNA levels of the miRNA processors Dicer, and DGCR8 using MDA-MB-231 cells treated with CNOT2 siRNA. In MDA-MB-231 breast cancer cells in which CNOT2 was knocked down by two siRNAs targeting different regions of the protein (Figure 1A), while the mRNA levels of Dicer and DGCR8 were attenuated (Figure 1B).
MiRNA expression following CNOT2 silencing
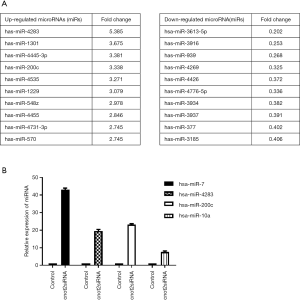
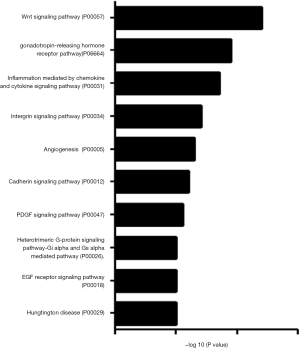
To determine whether CNOT2 regulates miRNA expression, we conducted gene expression profiling using an miRNA array and CNOT2 siRNA-treated MDA-MB-231 cells. The siRNA-induced silencing of CNOT2 upregulated the expression of 42 miRNAs, including has-miR-7, has-miR-4283, has-miR-10a, and has-miR-200c, whereas 47 genes, including has-miR-3916 and has-miR-3613-5p, were downregulated (Figure S1). The top 10 up- or down- expressed miRNAs from CNOT2 siRNA treated MDA-MB-23 cells are listed in Figure 2A. We also confirmed that has-miR-7, has-miR-4283, has-miR-200c, and has-miR-10a were upregulated in CNOT2 siRNA-treated cells (Figure 2B). The gene ontology annotations, according to the PANTHER PATHWAY database, included ten biological processes, including angiogenesis, Wnt signaling, and Huntington’s disease (Figure 3).
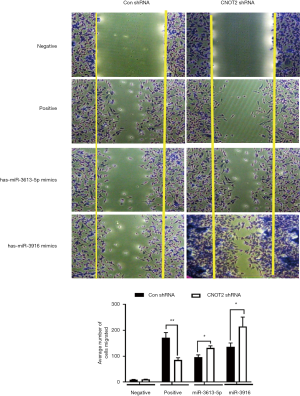
has-miR- 3613-5p and has-miR-3916 mimics rescued the inhibition of cell migration in CNOT2 depleted MDA-MD-231 cells
It was reported that silence of CNOT2 blocked migration of MDA-MB-231 cells (19). To see whether the miRNAs from miRNA array affect the migration, we performed the migration assay. We used has-miR-3613-5p and has-miR-3916 mimics which were the most downregulated from the miRNA array from CNOT2 siRNA treated MDA-MD-231 cells. The result showed that has-miR-3613-5p and has-miR-3916 rescued the inhibition of migration from CNOT2 shRNA stable cell lines (Figure 4).
Discussion
In the present study, a CNOT2 deficiency in MDA-MB-231 cells was found to modulate the expression of Dicer, and DGCR8, which are essential for miRNA processing. In addition, we examined miRNA expression in CNOT2 silenced MDA-MB231 cells. Overexpression by has-miR-3613-5p or has-miR-3916 mimic rescued the inhibition of migration from CNOT2 shRNA stable cell lines. Furthermore, we analyzed the pathways corresponding to the miRNA targeted genes to determine the function of CNOT2.
There is evidence that CCR4-NOT complex plays an important role in miRNA regulation (13). CNOT1 by interacting with MIF 4G- DEAD-box protein (DDX)6 contributes the repression of miRNA regulation (13). Also, GW182 repressed the function of miRNA by recruiting CNOT Complex (3). Here, we showed that CNOT2 regulates the mRNA level of Dicer, and DGCR8 which are miRNA processing complex and modulates miRNA expression in MDA-MB-231 cells.
It was reported that CNOT2 enhanced proliferation of MDA-MB-231 cells (19). Our study showed that has-miR-3613-5p and has-miR-3916 overexpression which were downregulated in miRNA array of CNOT2 depleted MDA-MB231 cells, rescued the inhibition of migration from CNOT2 shRNA stable cell lines. Thus, our data imply that has-miR- 3613-5p and has-miR-3916 might be important key factors in proliferation of CNOT2 in MDA-MB-231 cells.
Our analysis of miRNA within the PANTHER pathway database of miRNA targeted genes from CNOT2 depleted MDA-MB231 cells showed the following ontology: Huntington’s disease, Gi alpha and Gs alpha mediated pathway, and platelet-derived growth factor (PDGF) signaling pathway. Our data indicate that has-miR27-awas downregulated in CNOT2 siRNA-treated MDA-MB-231 cells. Chandrasekaran et al. (20) showed that has-miR27-a is a potential target in Huntington’s disease. has-miR-200c was upregulated while has-miR-21 was downregulated in the array produced from CNOT2 siRNA treated MDA-MB-231 cells, have been studied in relation to the PDGF signaling pathway. For instance, PDGF-BB treatment induced has-miR-200c in vascular smooth muscle cells (21). Furthermore, Wei et al. (22) showed that the inhibition of has-miR-21 expression by anti-has-miR-21 antibodies in LX-2 human hepatic stellate cells blocked PDGF-BB-stimulated LX-2 cell activation. The target genes of the miRNA in CNOT2 siRNA treated cells included SP-1 and BCAE-1, which are essential in the pathogenesis of neuroinflammation-associated diseases such as Alzheimer’s or Huntington’s disease (22).
In conclusion, the current study provides useful target genes and pathways for understanding the molecular mechanisms of CNOT2.

Acknowledgments
Funding: This was supported by
Footnote
Reporting Checklist: The author has completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-19-2821
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2821
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-19-2821
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2821). The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Collart MA, Panasenko OO. The Ccr4--not complex. Gene 2012;492:42-53. [Crossref] [PubMed]
- Zhang X, Virtanen A, Kleiman FE. To polyadenylate or to deadenylate: that is the question. Cell cycle 2010;9:4437-9. [Crossref] [PubMed]
- Chekulaeva M, Mathys H, Zipprich JT, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 2011;18:1218-26. [Crossref] [PubMed]
- Morita M, Oike Y, Nagashima T, et al. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/- mice. EMBO J 2011;30:4678-91. [Crossref] [PubMed]
- Ito K, Inoue T, Yokoyama K, et al. CNOT2 depletion disrupts and inhibits the CCR4-NOT deadenylase complex and induces apoptotic cell death. Genes Cells 2011;16:368-79. [Crossref] [PubMed]
- Shi J, Nelson MA. The cyclin-dependent kinase 11 interacts with NOT2. Biochem Biophys Res Commun 2005;334:1310-6. [Crossref] [PubMed]
- Chicoine J, Benoit P, Gamberi C, et al. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Developmental cell 2007;13:691-704. [Crossref] [PubMed]
- Aslam A, Mittal S, Koch F, et al. The Ccr4-NOT deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell 2009;20:3840-50. [Crossref] [PubMed]
- Takahashi A, Kikuguchi C, Morita M, et al. Involvement of CNOT3 in mitotic progression through inhibition of MAD1 expression. Biochemical and biophysical research communications 2012;419:268-73. [Crossref] [PubMed]
- Mittal S, Aslam A, Doidge R, et al. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol Biol Cell 2011;22:748-58. [Crossref] [PubMed]
- Zwartjes CG, Jayne S, van den Berg DL, et al. Repression of promoter activity by CNOT2, a subunit of the transcription regulatory Ccr4-not complex. J Biol Chem 2004;279:10848-54. [Crossref] [PubMed]
- Chen Y, Boland A, Kuzuoğlu-Öztürk D, et al. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 2014;54:737-50. [Crossref] [PubMed]
- Mathys H, Basquin J, Ozgur S, et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 2014;54:751-65. [Crossref] [PubMed]
- Fabian MR, Cieplak MK, Frank F, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 2011;18:1211-7. [Crossref] [PubMed]
- Behm-Ansmant I, Rehwinkel J, Doerks T, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 2006;20:1885-98. [Crossref] [PubMed]
- Castro D, Moreira M, Gouveia AM, et al. MicroRNAs in lung cancer. Oncotarget 2017;8:81679-85. [Crossref] [PubMed]
- Huang Y, Shen XJ, Zou Q, et al. Biological functions of microRNAs: a review. J Physiol Biochem 2011;67:129-39. [Crossref] [PubMed]
- Sohn EJ, Won G, Lee J, et al. Upregulation of miRNA3195 and miRNA374b Mediates the Anti-Angiogenic Properties of Melatonin in Hypoxic PC-3 Prostate Cancer Cells. J Cancer 2015;6:19-28. [Crossref] [PubMed]
- Sohn EJ, Jung DB, Lee H, et al. CNOT2 promotes proliferation and angiogenesis via VEGF signaling in MDA-MB-231 breast cancer cells. Cancer Lett 2018;412:88-98. [Crossref] [PubMed]
- Chandrasekaran S, Bonchev D. Network analysis of human post-mortem microarrays reveals novel genes, microRNAs, and mechanistic scenarios of potential importance in fighting huntington's disease. Comput Struct Biotechnol J 2016;14:117-30. [Crossref] [PubMed]
- Zheng B, Bernier M, Zhang XH, et al. miR-200c-SUMOylated KLF4 feedback loop acts as a switch in transcriptional programs that control VSMC proliferation. J Mol Cell Cardiol 2015;82:201-12. [Crossref] [PubMed]
- Wei J, Feng L, Li Z, et al. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed Pharmacother 2013;67:387-92. [Crossref] [PubMed]


