
Downregulation of ARID1A is correlated with poor prognosis in non-small cell lung cancer
Introduction
As the leading global cause of cancer-related death, lung cancer receives a considerable amount of attention from clinicians and researchers. Non-small cell lung cancer (NSCLC) comprises 80% of all lung cancer cases (1). Lung cancer patients may be treated through surgery, chemotherapy, radiotherapy, or best supportive care (BSC) (2). Despite the progress that has been made in understanding the biological characteristics of lung cancer and in generating new lung cancer therapies, the 5-year overall survival (OS) rate is suboptimal for lung cancer patients (3). Therefore, novel effective treatments are urgently needed to be developed. Numerous efforts have been made to improve the treatment efficacy for patients with advanced NSCLC. Identifying the biological and molecular diversity of lung cancer would help to improve traditional treatment modalities and support the development of novel targeted therapies.
AT-rich interaction domain 1A (ARID1A) is a tumor suppressor gene. Some studies have found that when its expression is dysregulated, ARID1A is involved in the progression of various tumors (4), such as the ovarian, endometrial epithelium (5), esophageal (6), breast (7), gastric (8), liver (9), pancreatic (10), bladder, and prostate cancer (11). However, there is little information regarding the role ARID1A plays in NSCLC, and the relationship between ARID1A and the clinicopathological features and prognosis of NSCLC patients has yet to be fully clarified. This study aimed to observe the expression of ARID1A in tumor specimens from human NSCLC patients and assess the clinicopathological significance of ARID1A in NSCLC, in an attempt to discover the potential impact of ARID1A on the occurrence of NSCLC.
We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2263).
Methods
Patient samples
Tumor specimens were collected from 108 patients (56 men and 52 women) with NSCLC who were treated in the Department of Thoracic Surgery at Jiangxi Chest Hospital between April, 2008, and April, 2009. The average age of the patients at the time of surgery was 61 years (range, 42–81 years) old. None of the patients had received chemotherapy or radiotherapy before surgery. Table 1 shows the patients’ characteristics and clinicopathological features. The 108 NSCLC cases were diagnosed by two experienced pathologists according to the histological classification criteria of the World Health Organization. All cases were staged and graded according to the 7th edition [2009] of the tumor, node, metastasis (TNM) classification system of the International Union Against Cancer (UICC) (12). The patients underwent radiological examination to determine whether there was distant metastasis. The 108 tumor specimens and 108 paracancerous normal lung (PCNL) tissues specimens were removed from the resected lung tissue at the time of surgery. The PCNL tissue was evaluated microscopically to determine the presence of normal cells and absence of atypical proliferative cells and was collected more than 5 cm from the edge of the tumor. The tissue specimens were excised within half an hour, and then divided into two parts: one part was frozen in liquid nitrogen for western blotting and the other part was fixed in 10% formalin for immunohistochemistry. In addition, 20 normal lung tissues were obtained from healthy living donors in our hospital. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Jiangxi Chest Hospital, Gan Xiong Lun Shen Zi [2016] No.4. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Table 1
| Clinical parameters | Total | ARID1A expression | χ2 | P valuea | ||
|---|---|---|---|---|---|---|
| High level | Low-moderate level | Absent | ||||
| Sex | 0.059 | 0.971 | ||||
| Male | 56 | 5 | 23 | 28 | ||
| Female | 52 | 4 | 22 | 26 | ||
| Age (years) | 3.069 | 0.216 | ||||
| ≥50 | 50 | 2 | 24 | 24 | ||
| <50 | 58 | 7 | 21 | 30 | ||
| Tumor size (cm) | 2.333 | 0.312 | ||||
| ≤3 | 62 | 3 | 27 | 32 | ||
| >3 | 46 | 6 | 18 | 22 | ||
| Histological type | 2.192 | 0.334 | ||||
| Squamous cell carcinoma | 51 | 4 | 25 | 22 | ||
| Adenocarcinoma | 57 | 5 | 20 | 32 | ||
| Smoking | 22.140 | <0.001* | ||||
| Yes | 60 | 6 | 36 | 18 | ||
| No | 48 | 3 | 9 | 36 | ||
| Differentiation | 10.800 | 0.005* | ||||
| Well/moderate | 70 | 8 | 35 | 27 | ||
| Poor | 38 | 1 | 10 | 27 | ||
| Lymphatic invasion | 8.756 | 0.013* | ||||
| Negative | 65 | 7 | 33 | 25 | ||
| Positive | 43 | 2 | 12 | 29 | ||
| Distant metastasis | 9.267 | 0.010* | ||||
| M0 | 71 | 7 | 36 | 28 | ||
| M1 | 37 | 2 | 9 | 26 | ||
| TNM stage | 13.768 | 0.001* | ||||
| Stage I/II | 63 | 7 | 34 | 22 | ||
| Stage III/IV | 45 | 2 | 11 | 32 | ||
a, statistical analyses were performed by the Pearson χ2 test; *, P<0.05 was considered significant. ARIDIA, AT-rich interaction domain 1a; NSCLC, non-small cell lung cancer.
Immunohistochemistry
Paraffin-embedded specimens were cut into 4-µm sections. Then, the protein expression of ARIDA1 was detected using immunohistochemistry with polyclonal rabbit anti-ARID1A antibody (HPA005456; Sigma-Aldrich, St Louis, MO, USA). The tissues were deparaffinized in xylene and rehydrated in alcohol. Following that, they were infiltrated in 3% hydrogen peroxide for 5 min. The sections were heated (100 °C) in 0.01 mol/L sodium citrate buffer (pH 6.0) for 30 min and rinsed 3 times for 5 min in phosphate buffered saline (PBS). The sections were incubated overnight at 4 °C with anti-ARID1A antibody diluted 1:200, and then incubated again with biotin-labeled secondary immunoglobulin at room temperature for 2 h. Finally, the color was developed with diaminobenzidine (DAB; DAKO, Glostrup, Denmark).
Three researchers used an Olympus BX51 microscope (Olympus, Center Valley, PA) to determine the immunohistochemical staining score (ISS) based on staining intensity and frequency. The staining intensity (SI) was classified as follows: 0 (negative); 1 (weak); 2 (moderate); and 3 (strong). The staining frequency (SF) was classified as 0 (no stained cells); 1 (1–25% of stained cells); 2 (25–50%); 3 (51–75%); and 4 (>75%). ISS values were obtained using the following calculation method: ISS = SI × SF. Immunoreactivity was classified according to ISS as: 0, negative (−); 1–4, weak (+); 5–8, moderate (++); and 9–12, strong (+++). The observers who evaluated the immunohistochemical staining results were kept blinded to the clinical characteristics and pathological parameters of the patients and each observers obtained similar results.
Western blotting
Total protein was extracted with lysate containing protease inhibitors, and western blotting was performed on 32 randomly selected paired NSCLC and PCNL tissue specimens. The total protein was separated using glycine-SDS-polyacrylamide gel and transferred to polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were blocked with TBST (tris-buffered saline containing 0.1% Tween-20) containing 5% skim milk for 2 h. Polyclonal rabbit anti-human ARID1A (Sigma-Aldrich, St. Louis, MO, USA) was diluted 1:200, rabbit anti-β-actin (Sigma-Aldrich, St. Louis, MO, USA) was used as an internal reference at a dilution of 1:1,000. Then, the PVDF membranes were incubated with the diluted antibody at 4 °C overnight. The next day, the membranes were washed 3 times in TBST for 5 min each, and then incubated with a peroxidase-conjugated goat anti-rabbit secondary antibody (1:1,000 dilution, Sigma-Aldrich) at room temperature for 2 h. The color was developed using a chemiluminescence system (Pierce, Rockford, IL, USA), and the film was displayed by enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ, USA).
Predictor and patient follow-up
After surgery, each patient was scheduled for follow-up examination, including physical examination, complete blood count, liver and kidney function examination and chest X-ray examination. The patients were follow-up once every 4 months for the first year, once every 6 months for the second year, and once a year thereafter. Outcome variables included tumor progression and cancer-related death. Event cases included patients whose tumors progressed or patients who died from metastasis cancer. Reviewed cases included patients for whom no endpoints were observed or patients for whom outcome status data were not available in the medical records. OS was measured from diagnosis to death or last follow-up (5 years). Patient deaths were determined by family report and verified by a review of the public records. The median follow-up of surviving patients was 31 months (range, 2–63 months).
Statistical analysis
Statistic data were analyzed using the SPSS 19.0 (SPSS, Chicago, IL, USA). Descriptive statistics data were expressed as mean ± SD (standard deviation) and significance was defined as P<0.05. The correlation between ARID1A expression and the clinicopathological features of the NSCLC patients was analyzed using the χ2 test. Spearman’s rank correlation analysis was carried out to examine the relationship between ARID1A expression and TNM stage. The Kaplan-Meier method was used to calculate survival, and differences in survival between groups were examined using log-rank analysis. Multivariate analysis of several independent prognostic factors was performed using Cox proportional hazards regression models.
Results
Protein expression level of ARID1A
First, the protein expression of ARID1A in 108 NSCLC tissues and PCNL tissues and 20 normal lung tissues by immunohistochemistry was observed. Positive ARID1A protein expression was detected in all normal lung tissues, 91.7% (99/108) of PCNL tissues (Figure 1A,B), and 50.0% (54/108) of NSCLC tissues. ARID1A protein levels were low in most of the NSCLC tissues (Figure 1C,D). Table 2 illustrates the results of immunohistochemistry. The ISS of the NSCLC tissues was significantly lower than that of the PCNL tissues or the normal control group (P<0.001). There was no statistical significance between the PCNL tissue and the normal control group (P>0.05).
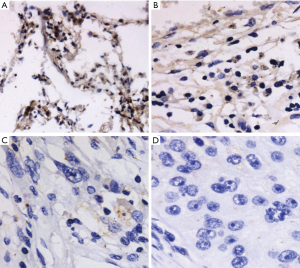
Table 2
| Clinical parameters | Number | ARID1A expression | Positive rate (%) | |||
|---|---|---|---|---|---|---|
| − | + | ++ | +++ | |||
| NSCLCa | 108 | 54 | 32 | 13 | 9 | 50.0 |
| Paracancerous tissuesb | 108 | 9 | 27 | 32 | 40 | 91.7 |
| lung normal tissuesc | 20 | 0 | 2 | 6 | 12 | 100.0 |
Statistical analyses were performed by the Pearson χ2 test; P<0.05 is considered significant. a/b: P<0.05 (χ2=60.201, P<0.001); a/c: P<0.05 (χ2=43.573, P<0.001); b/c: P>0.05 (χ2=5.534, P=0.137). ARIDIA, AT-rich interaction domain 1a; NSCLC, non-small cell lung cancer.
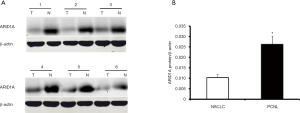
Then the 32 pairs of NSCLC tissues and PCNL tissues were randomly selected, and western blotting was performed to detect the protein expression of ARID1A. The results were similar to those of immunohistochemistry. The ARID1A protein expression value was expressed as the ratio of ARID1A/β-actin intensity. Compared with the PCNL tissues, 78.1% (25/32) of the NSCLC tissues had decreased ARID1A protein expression. Figure 2A illustrates the western blotting results of six typical cases. The mean protein expression level of ARID1A in the 32 NSCLC tissues was significantly lower than that in the PCNL tissues (P<0.05, Figure 2B).
The relationship between ARID1A expression and the clinicopathological features of NSCLC
Table 1 illustrates the relationship between the expression of ARID1A and the main clinicopathological features of the NSCLC patients. A significant correlation was found between the decrease in ARID1A expression and smoking (P<0.001), histological differentiation (P=0.005), lymphatic invasion (P=0.013), distant metastasis (P=0.010), and TNM stage (P=0.001). However, the expression of ARID1A in NSCLC tissues was not significantly correlated with age, gender, tumor size, or histological type (P>0.05).
Next, Spearman’s rank correlation analysis was used to explore the relationship between the expression of ARID1A in NSCLC tissues and TNM stage. Table 3 illustrates that the expression of ARID1A was negatively correlated with TNM stage; the more advanced the TNM stage, the lower the expression level of ARID1A in NSCLC tissue (rs=−0.366, P<0.001).
Table 3
| TNM stage | NEDD9 expression | Total | rs | P valuea | |||
|---|---|---|---|---|---|---|---|
| − | + | ++ | +++ | ||||
| I | 8 | 8 | 7 | 5 | 28 | −0.366 | <0.001* |
| II | 14 | 17 | 2 | 2 | 35 | ||
| III | 14 | 3 | 1 | 2 | 20 | ||
| IV | 18 | 4 | 3 | 0 | 25 | ||
| Total | 54 | 32 | 13 | 9 | 108 | – | – |
a, statistical analyses were performed by the Pearson χ2 test; *, P<0.05 was considered significant. ARIDIA, AT-rich interaction domain 1a; NSCLC, non-small cell lung cancer.
The expression of ARID1A and survival rates
Figure 3 illustrates the prognostic impact of ARID1A on OS. Based on the results of immunohistochemistry, the NSCLC patients were divided into the ARID1A negative and weak positive (− to +) groups and the ARID1A strong positive expression (+ to +++) groups. In the negative and weak positive (− to +) groups, there were 86 cases, out of whom 53 died; the 5-year OS rate was 53.6%. In the strong positive expression group (++ to +++) group, there were 22 cases, out of whom 10 died; the 5-year OS rate was 79.2%. The results showed that the lower the expression of ARID1A, the worse the prognosis of the patients (Figure 3A; χ2=134.448, P<0.001). Next, the survival time was analyzed based on ARID1A expression. The expression of ARID1A had no significant relationship with OS in stage I patients (Figure 3B; χ2=2.378, P=0.123). However, in patients with stages II to IV, the lower the ARID1A expression, the shorter the OS of the patients (Figure 3C, χ2=50.727, P<0.001; Figure 3D, χ2=103.971, P<0.001; Figure 3E, χ2=60.714, P<0.001). These results indicate that ARID1A may have a considerable impact on patients with advanced NSCLC.
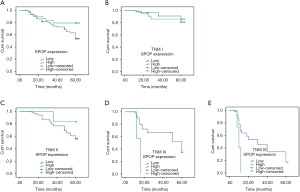
Table 4 illustrates the Cox regression analysis that was performed to investigate differences in response rates between patients with certain clinical factors. Univariate analysis revealed clinical variables, including tumor stage (P=0.035), lymphatic infiltration (P=0.044), tissue differentiation (P=0.033), tumor size (P=0.035), smoking (P=0.043), and ARID1A expression (P=0.011) were significantly correlated with OS. Multivariate analysis showed that the expression of ARID1A could be used as an independent predictor of OS in NSCLC patients.
Table 4
| Variables | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI | P valuea | HR | 95% CI | P valuea | ||
| Age | 1.456 | 0.898–2.639 | 0.172 | – | – | – | |
| Sex | 0.878 | 0.396–1.568 | 0.588 | – | – | – | |
| Histological type | 0.665 | 0.254–1.627 | 0.643 | – | – | – | |
| TNM stage | 2.021 | 1.050–3.887 | 0.035* | 2.357 | 0.695–3.895 | 0.064 | |
| Lymphatic invasion | 2.594 | 1.368–3.215 | 0.044* | 1.205 | 0.224–2.350 | 0.724 | |
| Differentiation | 2.050 | 1.002–3.886 | 0.033* | 1.996 | 0.014–3.635 | 0.442 | |
| Tumor size | 1.776 | 1.050–2.534 | 0.035* | 2.187 | 0.352–4.136 | 0.822 | |
| Smoking | 2.661 | 1.362–3.199 | 0.043* | 1.535 | 0.091–3.235 | 0.075 | |
| ARID1A expression | 2.889 | 1.224–5.127 | 0.011* | 2.357 | 1.315–4.895 | 0.024* | |
a, statistical analyses were performed by the Cox regression analysis; *, P<0.05 was considered significant.
Discussion
As the most common cause of cancer-related death, lung cancer is a disease of global concern. The most common cause of lung cancer is smoking, and is associated with environmental exposures (including secondhand smoke, pollution, and occupational carcinogens) as well as genetic susceptibility (13). NSCLC is a molecularly heterogeneous disease, and understanding its biological characteristics is essential for the treatment and prognosis of cancer. A number of molecular markers have been found to be strongly associated with poor prognosis in patients with NSCLC, such as over-expression of growth factor receptor (14) and low-expression of xeroderma pigmentosum group G (15). Compared with normal lung tissues, we found that the expression of ARID1A was lower in NSCLC tissues, and that low expression of ARID1A was closely related to poor prognosis of NSCLC patients, and our study provided a basis for ARID1A as a prognostic biomarker for NSCLC.
ARID1A can encode the BAF250A protein, which interacts with various other proteins, such as BRG1 and BRM adenosine triphosphatase to form SWI/SNF chromatin remodeling complex. This complex is crucial in regulating gene expression, cell development, cell differentiation, and tumor suppression. BAF250A is one of the subunits of the SWI/SNF complex (5). Moreover, the discovery that ARID1A is dysregulated in a variety of cancers has recently led to it becoming a research hotspot (5-11), and there is increasing evidence that ARID1A may function as a tumor suppressor (16). Dysregulation of ARID1A expression has been observed in many gynecological and non-gynecological tumors. We also found dysregulation of ARID1A expression in NSCLC. Immunohistochemistry and western blotting demonstrated that the expression of ARID1A in NSCLC tissues was decreased significantly compared with PCNL tissues and normal lung tissues. To be specific, the lower the protein expression level of ARID1A in NSCLC tissues, the worse the tumor differentiation. Furthermore, the expression level of ARID1A was also negatively correlated with some other important clinical parameters in the specimens, such as lymph node infiltration, distant metastasis, and TNM stage (all P<0.05). Therefore, ARID1A may be a potential diagnostic biomarker for NSCLC. These results also suggest that ARID1A may inhibit the cancer progression of NSCLC.
To date, the mechanism by which ARID1A expression is dysregulated and its role in carcinogenesis have remained unclear. Restoring wild-type expression levels of ARID1A can significantly suppress ovarian cancer cells and gastric cancer cells migration and invasion (5,17). Some studies involving leukemia cell lines have shown that knocking out ARID1A can increase resistance to Fas-mediated apoptosis (18). At the molecular level, Guan et al. found that the ARID1A/BRG1 complex binds to p53 and regulates several downstream effectors encoded by p21 and SMAD3 (5). Meanwhile, ARID1A can also regulate the activity of the PI3K pathway in endometrial cancer cell lines (19). When ARID1A is overexpressed, it can inhibit glioma cell proliferation as well as reduce the expression of p-AKT and p-S6K through the phosphatidylinositol 3-kinase pathway (20). In a recent report, Yokoyama et al. demonstrated that loss of ARID1A is related to chemoresistance in epithelial ovarian cancer (21). The finding implies that further analysis of ARID1A expression status may contribute to reducing the probability of resistance to platinum-based chemotherapy regimens, and may also improve sensitivity to PI3K/AKT inhibitors. PI3K/AKT inhibitors are likely to become a new way to overcome chemoresistance in patients with ARID1A-deficient tumors. However, the precise mechanism by which ARID1A is involved in NSCLC is not clear. Subsequent studies should focus on the precise mechanism of ARID1A in tumorigenesis, which may improve the diagnosis and treatment of ARID1A patients.
Our study found that the 5-year OS rates of the ARID1A negative and weak-positive (− to +) groups were significantly higher than that of the ARID1A strongly positive (++ to +++) group. Spearman’s rank correlation analysis was used and found that the lower the ARID1A in NSCLC, the more advanced the clinical TNM stage. The results of univariate survival analysis showed that decreased ARID1A expression was significantly associated with prognosis. The results of multivariate survival analysis suggested that the expression of ARID1A could be used as an independent factor for predicting the prognosis of NSCLC; when ARID1A expression is low, it may indicate a poor prognosis for patients. Moreover, Spearman’s rank correlation analysis indicated that the expression of ARID1A in NSCLC was negatively correlated with TNM stage; that is, the lower the level of ARID1A expression the later the clinical TNM stage. These results suggest that ARID1A plays a role in the development and progression of NSCLC. However, other findings suggest a more complex role for ARID1A in tumorigenesis. For example, some SWI/SNF components being oncogenic in some cases (22,23); ARID1A expression levels are elevated in most liver cancer tumors compared with adjacent non-cancerous tissues (24); and ARID1A inactivation promotes epithelial differentiation and prolongs survival in APC and PTEN-deficient mouse ovarian cancer models (25). Therefore, the precise mechanism of ARID1A in NSCLC warrants further investigation in order to provide new ideas for the prognosis or treatment of NSCLC.
Conclusions
Abnormal ARID1A expression is common in NSCLC. In this study, we observed that ARID1A expression was significantly decreased in NSCLC tissues compared to PCNL tissues. Compared with the ARID1A strong-positive group, the OS of the ARID1A negative/weak-positive group was significantly shorter. In summary, our results suggest that the down-regulation of ARID1A is correlated with poor prognosis in NSCLC. ARID1A expression will play an important role as a predictive biomarker for poor survival.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2263
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2263
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2263). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of Jiangxi Chest Hospital, Gan Xiong Lun Shen Zi (2016) No.4. Because of the retrospective nature of the research, the requirement for informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Herbst RS, Heymach JV, Lippman SM. Lung Cancer. N Engl J Med 2008;359:1367-80. [Crossref] [PubMed]
- Zaman A, Bivona TG. Emerging application of genomics-guided therapeutics in personalized lung cancer treatment. Ann Transl Med 2018;6:160. [Crossref] [PubMed]
- Subramanian MP, Puri V. Neoadjuvant vs. adjuvant chemotherapy in locally advanced non-small cell lung cancer—is timing everything? J Thorac Dis 2019;11:5674-6. [Crossref] [PubMed]
- Jones S, Li M, Parsons DW, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat 2012;33:100-3. [Crossref] [PubMed]
- Guan B, Wang TL, Shih I. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res 2011;71:6718-27. [Crossref] [PubMed]
- Agrawal N, Jiao Y, Bettegowda C, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov 2012;2:899-905. [Crossref] [PubMed]
- Cornen S, Adelaide J, Bertucci F, et al. Mutations and deletions of ARID1A in breast tumors. Oncogene 2012;31:4255-6. [Crossref] [PubMed]
- Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet 2012;44:570-4. [Crossref] [PubMed]
- Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012;44:760-4. [Crossref] [PubMed]
- Shain AH, Giacomini CP, Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci U S A 2012;109:E252-9. [Crossref] [PubMed]
- Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 2011;43:875-8. [Crossref] [PubMed]
- Zhong X, Li M, Nie B, et al. Overexpression of RACK1 and CD147 associated with poor prognosis in stage T1 pulmonary adenocarcinoma. Ann Surg Oncol 2013;20:1044-52. [Crossref] [PubMed]
- Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e1S-e29S.
- Selvaggi G, Novello S, Torri V, et al. Epidermal growth factor receptor overexpression correlates with a poor prognosis in completely resected non-small-cell lung cancer. Ann Oncol 2004;15:28-32. [Crossref] [PubMed]
- Bartolucci R, Wei J, Sanchez JJ, et al. XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung Cancer 2009;10:47-52. [Crossref] [PubMed]
- Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov 2013;3:35-43. [Crossref] [PubMed]
- Yan HB, Wang XF, Zhang Q, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis 2014;35:867-76. [Crossref] [PubMed]
- Luo B, Cheung HW, Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 2008;105:20380-5. [Crossref] [PubMed]
- Liang H, Cheung LW, Li J, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res 2012;22:2120-9. [Crossref] [PubMed]
- Zeng Y, Liu Z, Yang J, et al. ARID1A is a tumour suppressor and inhibits glioma cell proliferation via the PI3K pathway. Head Neck Oncol 2013;5:6.
- Yokoyama Y, Matsushita Y, Shigeto T, et al. Decreased ARID1A expression is correlated with chemoresistance in epithelial ovarian cancer. J Gynecol Oncol 2014;25:58-63. [Crossref] [PubMed]
- Jubierre L, Soriano A, Planells-Ferrer L, et al. BRG1/SMARCA4 is essential for neuroblastoma cell viability through modulation of cell death and survival pathways. Oncogene 2016;35:5179-90. [Crossref] [PubMed]
- Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013;153:71-85. [Crossref] [PubMed]
- Zhao J, Chen J, Lin H, et al. The Clinicopathologic Significance of BAF250a (ARID1A) Expression in Hepatocellular Carcinoma. Pathol Oncol Res 2016;22:453-9. [Crossref] [PubMed]
- Zhai Y, Kuick R, Tipton C, et al. Arid1a inactivation in an Apc- and Pten-defective mouse ovarian cancer model enhances epithelial differentiation and prolongs survival. J Pathol 2016;238:21-30. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


