
Application of metagenomic next-generation sequencing technology for difficult lung lesions in patients with haematological diseases
Introduction
The treatment of haematological malignancies includes chemotherapy, radiotherapy, bone marrow transplantation, biological therapy, target gene therapy, and other treatments. Chemotherapy in patients with haematological diseases leads to myelosuppression, a decrease in the number of neutrophils, and reduced immune function, resulting in more lung infections by various unknown pathogens. Given long-term use of broad-spectrum antibiotics, patients with haematological diseases are highly susceptible to infections with viruses, bacteria, invasive fungi, and even various resistant bacteria (1). The patient’s condition is worse, the cost is higher, and the precise treatment of the target pathogen is poor. The diagnostic interventional pulmonology procedure rapid on-site evaluation of cytology (ROSE) is a real-time cytological examination technique that accompanies sequential sampling. A small part of every tissue specimen sampled from the target lesion is smeared on a slide without significant loss of tissue material. Metagenomic next-generation sequencing (mNGS) technology does not rely on traditional microbial culture but instead directly performs high-throughput sequencing of nucleic acids in clinical samples and then compares the obtained sequence information with databases (2). Identification of microbial species can lead to quick and objective detection of more pathogenic microorganisms in clinical samples, especially for the diagnosis of critically ill patients and those with difficult infections (3,4). The purpose of this study was to evaluate the diagnostic value of combined virtual bronchoscopic navigation (Direct Path), radial endobronchial ultrasound with guide-sheath (EBUS), ultrathin bronchoscopy, ROSE, and mNGS for difficult lung lesions in patients with haematological diseases. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-604).
Methods
Patients
A total of 81 patients underwent bronchoscopy at the respiratory endoscopy centre of Tianjin Medical University General Hospital from July 2018 to January 2019. These patients were admitted to the Institute of Hematology and Blood Disease Hospital, Chinese Academy of Medical Sciences, and had completed more than one round of chemotherapy. The diagnosis of patients with complicated lung lesions is not clear, and treatment with multiple antibiotics is not ideal. A total of 15 cases had acute lymphoblastic leukaemia, 27 had acute non-lymphocytic leukaemia, 21 had myelodysplastic syndrome, 9 had lymphoma, and 9 had aplastic anaemia. Eighteen of these haematological patients underwent haematopoietic stem cell transplantation. The median age of the patients was 42.3 years (range, 16–63 years). All patients were fully informed of the test content and risks and signed an informed consent form before surgery. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Committee of Tianjin Medical University General Hospital, China approved this study (No. IRB2018-133-01). An electrocardiogram (ECG), routine blood examination, C-reactive protein (CRP), coagulation, blood biochemistry, hepatitis B, human immunodeficiency virus (HIV), syphilis, procalcitonin quantitation (PCT), serum 1,3-β-D-glucan test (BG test), galactomannan test (GM test), sputum smear microscopy, and sputum culture were performed before surgery.
Specimen collection
Bronchoscopy [routine bronchoscopy: BF-F260, 5.5-mm outer diameter of the tip (Olympus, Tokyo, Japan) and ultrathin bronchoscopy: BF-P-260F, 4-mm outer diameter of the tip (Olympus)] to collect samples, including transbronchial lung biopsy (TBLB) and BAL, were performed in subjects. For patients with peripheral lesions that made obtaining specimens difficult, ultrathin bronchoscopy under radial EBUS and virtual bronchial imaging using virtual navigation were guided along the route displayed on the virtual image to direct the location to or near to the lesion. The operating instrument was placed, and the specimen was collected. The combination with ROSE yielded 6–10 diagnostically valuable lung tissue specimens and alveolar lavage fluid. The same physician collected the above specimens. A small amount of lung tissue specimen was collected, and a circle approximately 1 cm in diameter was smeared on a sterile slide using a disposable 2.5–5.0-mL syringe needle for sterile cytology. Then, the slides were immediately placed in Diff Quik (DQ) AB staining solution (Zhuhai Besso Biological Company, China) and rapidly stained sequentially in solution A (30 seconds) and solution B (40 seconds). The lung tissue specimens were sent for pathology. The affected bronchopulmonary segment was lavaged 4 times with 60–80 mL of sterile saline at 37 °C. A total of 5–15 mL of alveolar lavage fluid was collected for DNA extraction and stored at −80 °C.
NGS procedure for sample
Zero point five to 3 mL sample from patient was collected according to standard procedures. One point five mL microcentrifuge tube with 0.5 mL sample and 1 g 0.5 mm glass bead were attached to a horizontal platform on a vortex mixer and agitated vigorously at 2,800–3,200 RPM for 30 min. Zero point three mL sample was separated into a new 1.5 mL microcentrifuge tube and DNA was extracted using the TIANamp Micro DNA Kit (DP316, TIANGEN BIOTECH) according to the manufacturer’s recommendation. Then, DNA libraries were constructed through DNA-fragmentation, end-repair, adapter-ligation and PCR amplification. Agilent 2100 was used for quality control of the DNA libraries. Quality qualified libraries were sequenced by BGISEQ-50 platform.
Statistical analysis
The data were statistically analysed using the SPSS 17.0 software. The count data were represented by n (%). Differences were considered significant if the P values were <0.05. Two-by-two contingency tables were derived to determine sensitivity (Se), specificity (Sp), positive predictive value (PPV), and negative predictive value (NPV). All statistics are reported as absolute values with 95% confidence intervals (95% CIs). Comparative analyses were conducted by using the McNemar test.
Results
Laboratory test results
The characteristics of patients with haematological diseases are shown in Table 1. Laboratory test results for the patients with haematological diseases are presented in Table 2. The patients’ platelets were significantly lower than the normal range.
Table 1
| Characteristics | Patients (N=81) |
|---|---|
| Sex | |
| Female | 33 [41] |
| Age, years | |
| ≤20 | 9 [11] |
| 21–60 | 69 [85] |
| >60 | 3 [4] |
| Initial diagnosis | |
| HAP | 30 [37] |
| TB | 9 [11] |
| Aspergillus | 6 [7] |
| PCP | 9 [11] |
| Non-specific | 18 [22] |
| Uncertain | 9 [11] |
All values are indicated as the number of cases [percentage]. HAP, hospital acquired pneumonia; TB, tuberculosis; PCP, Pneumocystis Jirovecii pneumonia; non-specific, nonspecific inflammation.
Table 2
| Laboratory test | Patients (N=81) |
|---|---|
| WBC (×109/L) | 6.44±3.06 |
| PLT (×109/L) | 61.10±32.98 |
| CRP (mg/dL) | 55.98±27.85 |
| PCT (ng/mL) | 1.39±1.27 |
| GM test | 0.68±0.64 |
| BG test (pg/mL) | 68.79±4.27 |
All values are indicated as a mean and standard deviation. WBC, white blood cell count; PLT, platelet count; CRP, C-reactive protein; PCT, procalcitonin quantitation; GM test, galactomannan test; BG test, 1,3-β-D-glucan test.
Lung biopsy cytology results
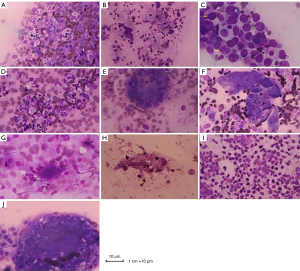
Six cases were consistent with purulent infection (Figure 1A), and 6 cases were consistent with general inflammatory changes (Figure 1B). In 11 cases, the distal airway epithelial cells proliferated and hypertrophied into a giant cell reaction (yellow arrow), and the nuclei generally were greater than twice the diameter of those of the red blood cells. Deep staining showed inclusion bodies (green arrow) in the nuclei, which could indicate viral infection (Figure 1C). In one case, the main infiltrating inflammatory cell type was monocytes, which could represent infection with mycoplasma and Chlamydia (Figure 1D). Twelve cases were consistent with granulomatous inflammation (Figure 1E). In 16 cases, foam-like macrophages aggregated into clusters, which was a typical feature of Mason’s bodies; additionally, alveolar septal thickening was visualized, which was consistent with the organization (Figure 1F). Twelve cases showed more activated lymphocytes, and the background of fluffy cotton-like protein was deeply stained with small spots indicative of a trophozoite, which might be consistent with Pneumocystis jirovecii pneumonia (PCP) (Figure 1G). In eight cases, tubular hyphae with diameters of 2 to 7 µm were observed, and an acute angle of approximately 45° formed at the hyphae (Figure 1H). A large number of eosinophils were observed in six cases, which were highly suggestive to fungal infections. However, no intact hyphae were noted using ROSE, and broken hyphae structures were observed (Figure 1I). In three cases, many reactive lymphocytes and round cryptococcal capsuled cysts that were 4–6 micron in size were observed in the cytoplasm of multinucleated giant cells (Figure 1J).
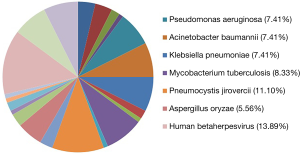
Distribution and composition ratios of mNGS detection of pathogenic microorganisms in the patients’ lung tissues and alveolar lavage fluid (%)
The mNGS did not detect pathogens in lung tissues from nine patients. In the remaining patients, prokaryotic microorganisms accounted for 44.44% of the detected sequences, eukaryotic microorganisms accounted for 26.85%, and viruses accounted for 28.70%. The main pathogens were cytomegalovirus, Pneumocystis jirovecii, and Mycobacterium tuberculosis. mNGS of the alveolar lavage fluid revealed pathogenic microorganisms in 44.44% of the samples, including eukaryotic microorganisms in 27.35% and viruses in 28.21%. During collection of the patient specimens and bronchoalveolar lavage fluid (BALF), mNGS detected oral microbes and bacteria; however, the combined clinical analysis results were not within the statistical range (Table 3). The percentage distributions of pathogenic microorganisms in lung tissue detected by mNGS are shown in Figure 2.
Table 3
| Pathogenic microorganism | TBLB | BALF | |||
|---|---|---|---|---|---|
| Number of strains | Composition ratio | Number of strains | Composition ratio | ||
| Prokaryotic microorganism | 48 | 44.44 | 52 | 44.44 | |
| Actinomyces odontolyticus | 4 | 3.70 | 5 | 4.27 | |
| Enterococcus faecium | 4 | 3.70 | 2 | 1.71 | |
| Nocardia cyriacigeorgica | 2 | 1.85 | 2 | 1.71 | |
| Nocardia africana | 1 | 0.93 | 1 | 0.85 | |
| Pseudomonas aeruginosa | 8 | 7.41 | 11 | 9.40 | |
| Acinetobacter baumannii | 8 | 7.41 | 9 | 7.69 | |
| Klebsiella pneumoniae | 8 | 7.41 | 10 | 8.55 | |
| Enterobacter hormaechei | 2 | 1.85 | 2 | 1.71 | |
| Chlamydia psittaci | 1 | 0.93 | 0 | 0 | |
| Mycobacterium tuberculosis | 9 | 8.33 | 9 | 7.69 | |
| Mycobacterium kansasii | 1 | 0.93 | 1 | 0.85 | |
| Eukaryotic microorganism | 29 | 26.85 | 32 | 27.35 | |
| Pneumocystis jirovercii | 12 | 11.10 | 12 | 10.26 | |
| Cryptococcus neoformans | 3 | 2.78 | 2 | 1.71 | |
| Aspergillus oryzae | 6 | 5.56 | 5 | 4.27 | |
| Aspergillus fumigatus | 3 | 2.78 | 6 | 5.13 | |
| Aspergillus niger | 1 | 0.93 | 3 | 2.56 | |
| Aspergillus flavus | 2 | 1.85 | 2 | 1.71 | |
| Aspergillus tubingensis | 1 | 0.93 | 1 | 0.85 | |
| Rhizopus delemar | 1 | 0.93 | 1 | 0.85 | |
| Virus | 31 | 28.70 | 33 | 28.21 | |
| Human betaherpesvirus 5 | 15 | 13.89 | 18 | 15.38 | |
| Human gammaherpesvirus 4 | 8 | 7.41 | 9 | 7.69 | |
| Human alphaherpesvirus 1 | 8 | 7.41 | 6 | 5.13 | |
| Total | 108 | 100.00 | 117 | 100.00 | |
All values are indicated as the number of strains and percentage. mNGS, metagenomic next-generation sequencing; TBLB, transbronchial lung biopsy; BALF, bronchoalveolar lavage fluid.
Combination of clinical final outcomes, laboratory examination results, pathological findings, ROSE, and mNGS comprehensive diagnostic results
Eight cases were graft-versus-host disease (GVHD), one case was idiopathic pneumonia syndrome (IPS), and two cases were delayed pulmonary toxicity syndrome (DPTS). Fourteen cases were pulmonary aspergillosis, 12 cases were PCP, three cases were pulmonary cryptococcosis, ten cases were tuberculosis, one case was nontuberculous mycobacteria (NTM), 15 cases were cytomegalovirus pneumonia, six cases were Pseudomonas aeruginosa pneumonia, six cases were drug-resistant Klebsiella pneumoniae, one case was Chlamydia pneumoniae, and three cases were pulmonary nocardiosis.
Comparison of the Se, Sp, PPV and NPV of mNGS of pathogenic microorganisms in the TBLB and BALF
The Se and Sp of the TBLB mNGS were 91.67% and 66.66%, respectively, and the PPV and NPV were 96.65% and 50.00%, respectively. The Se and Sp of mNGS of the BALF were 76.19% and 74.29%, respectively, and the PPV and NPV were 72.72% and 76.67%, respectively. The pathogenic microbial Se of lung tissue was increased compared with BALF. The Sp of the BALF mNGS was better than that of lung tissue. The mNGS detection results for the TBLB were accurate, whereas those obtained for the BALF were broader. Thus, the two methods have unique advantages.
Discussion
Pulmonary complications in patients with haematological diseases can be divided into two categories: infectious pulmonary complications caused by bacteria, fungi, viruses, tuberculosis and other pathogens and non-infectious pulmonary complications, such as pulmonary edema, diffuse alveolar haemorrhage (DAH), IPS (5), DPTS, GVHD, bronchiolitis obliterans (BO) with organizing pneumonia (BOOP), and BO (6). In the past, infectious pulmonary complications with unclear diagnoses were poorly targeted by anti-infective treatment, resulting in poor infection control and delays in treatment of patients’ diseases. Non-infectious pulmonary complications in patients with haematological diseases are often treated with a variety of antibiotics, resulting in poor treatment due not only to a waste of medical expenses but also delayed treatment of the disease.
Infections associated with chemotherapy in patients with haematological diseases are a leading cause of treatment failure and an important cause of death (7). Haematological diseases complicated with pulmonary infection mostly include hospital-acquired pneumonia (HAP), the causes of which are as follows: (I) destruction of the human immune system by basic blood diseases (8); (II) myelosuppression caused by chemotherapeutic drugs (9); (III) use of a large amount of glucocorticoids and immunosuppressive agents; (IV) long-term use of a variety of broad-spectrum antibiotics that causes dysbacteriosis of the flora, thereby making these patients a high-risk group for invasive fungal infections. The common pathogens of HAP complicated by blood diseases include extremely high incidences of bacteria, fungi, tuberculosis, rare pathogens, and drug-resistant bacteria. These patients are prone to developing severe pneumonia; additionally, infection is often present at increased levels, the disease progresses rapidly, the mortality rate and costs are high, and the accuracy of detection of the target pathogen is poor.
With the continuous development of molecular biological technology, human research on diseases is no longer limited to a single genetic locus but instead focuses on the whole genome. NGS technology is an open analysis and diagnostic system. Currently, greater than 8,000 pathogens are known, including more than 3,000 types of bacteria, 4,000 types of viruses, 200 types of fungi and 140 types of parasites. Diagnosis of rare pathogenic microbial infections provides an effective technical approach (10). mNGS technology represented by Illumina MiSeq and HiSeq has developed rapidly (11). mNGS has greatly improved the sequencing throughput, shortened the sequencing time, reduced the cost of sequencing, and ushered in a revolutionary breakthrough in the diagnosis of infectious diseases. ROSE is a rapid cytological interpretation technique that is accompanied by a material acquisition process (12). The content of the interpretation includes cell morphology, classification, counting, composition ratio, arrangement, correlation, background, and foreign object analysis. Patients with haematological diseases treated via chemotherapy, which causes bone marrow suppression and thrombocytopenia, are prone to bleeding. Conventional TBLB has a low diagnostic rate for peripheral lung lesions, whereas CT-guided percutaneous lung biopsy has increased risks of bleeding and pneumothorax. Asano showed that the diagnostic accuracy of virtual endoscopy combined with ultrathin bronchoscopy for peripheral lesions with a diameter ≤30 mm was 67.1% (13). At present, for virtual bronchoscopic navigation (Direct Path), radial EBUS, and ultrathin bronchoscopy combined with mNGS technology, the targets are collected for rapid on-site cytological evaluation to reduce the bleeding risk and patient pain and trauma. The use of mNGS to sequence pathogenic microorganisms has greatly improved the diagnostic rate of patients and thus better guides their treatment. mNGS is a critical key technology for achieving accurate diagnoses of infectious diseases (14,15).
In this study, the diagnosis of infectious pneumonia mainly included cytomegalovirus pneumonia, PCP, pulmonary aspergillosis and tuberculosis. The use of ROSE for cytology of lung tissue biopsies revealed results that were consistent with viral infection and granulomatous inflammation with the corresponding hyphae, pathogen spores, cysts, and trophozoites. Patients with haematological diseases are highly suspected of having PCP. However, the BALF examination was negative. Alveolar lavage fluid was examined by hexamine silver and PAS staining, but the results were negative. Finally, a BALF review by hexamine silver staining and microscopic examination found suspected Pneumocystis jirovecii. If the patient’s pathogen cannot be found, then the disease cannot be diagnosed, and the patient’s therapeutic medication cannot reach the therapeutic dose, resulting in the failure of the patient's overall treatment. However, lung tissue and BALF mNGS detected Pneumocystis jirovecii. The number of pathogens detected in lung tissue was significantly increased compared with that in the alveolar lavage fluid, thereby allowing for the early diagnosis and precise treatment of the disease. In this study, six patients with ROSE results showed a large number of eosinophils, which were highly suggestive of fungal infections. However, no intact hyphae were identified by ROSE, and broken hyphae structures were observed. Pathological examination of the patient indicated the presence of mould mycelia, suggesting that the patient had a fungal infection. However, mNGS analysis revealed Aspergillus oryzae and other pathogens. Thus, NGS technology can accurately detect the genera of eukaryotic microorganisms.
This study shows that BALF can be used to detect the range of pathogenic microorganisms in patients following mNGS of pathogenic microorganisms, which has advantages for patients for whom specimens cannot be accurately obtained, thereby narrowing the scope of the diagnosis. In contrast, lung biopsy accurately collects lesions for mNGS of microorganisms to obtain a precise diagnosis. However, when obtaining a specimen is difficult, the disease needs to be diagnosed by BALF. Lung tissue specimens obtained by bronchoscopic lung biopsy (TBLB) represent one or more collection points combined with ROSE to obtain representative lesions. The specimen contains high concentrations of pathogens, and the technique exhibits good Se and Sp. If a representative lesion is obtained, it is easy to distinguish between infection and colonization. BALF represents one or more sampling groups in the lungs. If the lavage is representative of the lesion, it may contain a certain concentration of the pathogen. However, because BALF contains more bacteria in the oral cavity and main airway, it is more polluted and has a higher microbial load. Thus, its Se is lower than TBLB. TBLB has higher Se for mNGS, whereas BALF has higher Sp. Therefore, both TBLB and BAL have different advantages. In this study, one patient was highly suspected of having Klebsiella pneumoniae pneumonia based on a combination of their medical history, laboratory tests, and multiple sputum cultures. mNGS of the BALF showed an increased presence of Klebsiella pneumoniae, but compared with the alveolar lavage fluid, the number of sequences in lung tissue detected by mNGS was significantly reduced because the patient was in critical condition and on mechanical ventilation. The patient’s lung tissue was taken from the respiratory intermediate care unit (RICU). The target specimen was not taken under the guidance of ROSE, which led to error of the number of sequences detected in lung tissue by mNGS.
In this study, some patients had non-infectious pulmonary complications, and the clinical and therapeutic outcomes were diagnosed as GVHD, IPS and DPTS. GVHD is a serious complication after transplantation in patients with haematological diseases and is the main cause of the increased prevalence and non-recurring mortality of infections in patients (16). The organ with the longest GVHD involvement is the skin, which can affect lung tissue; however, the incidence is low. The definition of lung chronic GVHD (cGVHD) in the Seattle standard is vague, and lung lesions are difficult to distinguish from other diseases, such as infection (17). The National Institutes of Health’s (NIH’s) Consensus provides a more detailed definition of lung cGVHD. BO demonstrated by pulmonary function and radioactivity requires the characteristic features of cGVHD in at least one other organ (18). Bronchiolitis is associated with the manifestation of GVHD in lung tissue. BO is the only diagnostic clinical manifestation of GVHD. In this study, ROSE found aggregation of foam-like macrophages in groups, and some macrophages evolved into tissue and epithelial cells. More fibroblasts appeared, some of which had evolved into fibrocytes. The combination of the patient’s medical history and the lack of detection of the main pathogenic bacteria by mNGS led us to consider the patient as having pulmonary GVHD, which provided a new diagnostic approach for the diagnosis of lung transplant-versus-host disease. However, the patient’s final clinical outcome was diagnosed as IPS and DPTS; thus, the microbes detected by mNGS were not real pathogenic microorganisms.
mNGS has a low detection rate of intracellular bacteria and thick-walled microorganisms. Therefore, even in the test report, the sequence number of this microorganism is not high. When background bacteria, contaminating bacteria, and colonizing bacteria are excluded, the possibility of pathogenic microorganisms is considered. With the optimization and improvement of detection technology, the Se of mNGS in the diagnosis of intracellular bacterial infections is increasing. mNGS can sequence DNA and RNA simultaneously. Each sample is introduced with an internal reference, and each batch of the sample is introduced with a negative quality control and environmental quality control to control pollution to the greatest extent. The genome of some viruses is RNA, which cannot be detected by DNA library construction. In addition, RNA and DNA sequencing results can confirm and complement each other. In addition, RNA abundance is positively correlated with gene transcription activity. Detection of RNA can identify dead and live bacteria, distinguishing between past and current infections. Therefore, DNA + RNA sequencing has multiple advantages over simple DNA sequencing, especially in children’s infections. However, human-derived RNA exhibits increased abundance and complexity compared with DNA, and RNA is easily degraded and has more stringent requirements for sample transportation and storage. Achieving stable and effective simultaneous detection of DNA and RNA will guarantee improvements in the clinical detection rate and accuracy of mNGS. Combining clinical and patient economic issues in this study, DNA testing is exclusively currently performed on patient lung tissues. Currently, RNA sequencing may be particularly useful in clinical cases where the pathogen is only transient and may identify whether an acute disease is an infectious disease. However, RNA is easily degraded, and the requirements for detection are high. When interpreting the RNA pathogen report, it is necessary to consider specimen transportation and laboratory conditions, and currently there are still some difficulties in clinical detection. At present, due to the low coverage of related resistance genes, it is difficult to detect related resistance genes with high Se. With the advancement of technology, mNGS detection technology with high coverage of drug resistance genes will gradually be developed.
The results of this study indicate that use of combined virtual bronchoscopic navigation (Direct Path), radial EBUS, ultrathin bronchoscopy, ROSE and mNGS technology has high clinical application value for patients with haematological diseases and other immunosuppressive hosts with serious infections, severe lung infections and other diseases. Use of these technologies in combination with traditional methods can improve the Se and Sp of pathogen diagnosis. For infectious pulmonary complications, mNGS detection can reduce the unnecessary use of antibiotics and related medical expenses and result in better disease treatment (19). The shortcomings of this study are as follows: (I) this study is a single-centre study; the sample size in this study and the scope of the examination area are small and thus are not sufficient to reflect the overall outline of the disease; (II) background bacterial contamination may be detected by mNGS; (III) host individual heterogeneity, an insufficiently high pathogen microbial load and different stages of the disease have certain effects on detection; (IV) the sequencing platform, data analysis, and report interpretation have key impacts on the sequencing results.
Acknowledgments
The authors thank the patients for participating and donating samples to make this research possible, acknowledgment the support of endoscopy center of the General Hospital of Tianjin Medical University, and Institute of Hematology and Blood Disease Hospital, Chinese Academy of Medical Sciences professors provide cases, and Tianjin Academy of Traditional Chinese Medicine Affiliated Hospital professors collect data.
Funding: This work was supported by the grants from
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-604
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-604
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-604). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy of integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The trial was approved by the Ethics Committee of Tianjin Medical University General Hospital, China (No. IRB2018-133-01). Written informed consent was obtained from all patients with haematological diseases.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dallas J, Kollef M. Severe hospital-acquired pneumonia: a review for clinicians. Curr Infect Dis Rep 2009;11:349-56. [Crossref] [PubMed]
- Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet 2019;20:341-55. [Crossref] [PubMed]
- Didelot X, Bowden R, Wilson DJ, et al. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 2012;13:601-12. [Crossref] [PubMed]
- Westblade LF, van Belkum A, Grundhoff A, et al. Role of Clinicogenomics in Infectious Disease Diagnostics and Public Health Microbiology. J Clin Microbiol 2016;54:1686-93. [Crossref] [PubMed]
- Afessa B, Litzow MR, Tefferi A. Bronchiolitis obliterans and other late onset non-infectious pulmonary complications in hematopoietic stem cell transplantation. Bone Marrow Transplant 2001;28:425-34. [Crossref] [PubMed]
- Freudenberger TD, Madtes DK, Curtis JR, et al. Association between acute and chronic graft-versus-host disease and bronchiolitis obliterans organizing pneumonia in recipients of hematopoietic stem cell transplants. Blood 2003;102:3822-8. [Crossref] [PubMed]
- Garcia JB, Lei X, Wierda W, et al. Pneumonia during remission induction chemotherapy in patients with acute leukemia. Ann Am Thorac Soc 2013;10:432-40. [Crossref] [PubMed]
- Kollef MH, Sherman G, Ward S, et al. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 1999;115:462-74. [Crossref] [PubMed]
- Yoo JH, Choi SM, Lee DG, et al. Prognostic factors influencing infection-related mortality in patients with acute leukemia in Korea. J Korean Med Sci 2005;20:31-5. [Crossref] [PubMed]
- Jerome H, Taylor C, Sreenu VB, et al. Metagenomic next-generation sequencing aids the diagnosis of viral infections in febrile returning travellers. J Infect 2019;79:383-8. [Crossref] [PubMed]
- Lazarevic V, Whiteson K, Huse S, et al. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods 2009;79:266-71. [Crossref] [PubMed]
- Ganc RL, Carbonari AP, Colaiacovo R, et al. Rapid on-site cytopathological examination (ROSE) performed by endosonagraphers and its improvement in the diagnosis of pancreatic solid lesions. Acta Cir Bras 2015;30:503-8. [Crossref] [PubMed]
- Asano F, Shinagawa N, Ishida T, et al. Virtual bronchoscopic navigation combined with ultrathin bronchoscopy. A randomized clinical trial. Am J Respir Crit Care Med 2013;188:327-33. [Crossref] [PubMed]
- Bayer R, Galea S. Public Health in the Precision-Medicine Era. N Engl J Med 2015;373:499-501. [Crossref] [PubMed]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793-5. [Crossref] [PubMed]
- Lee SJ, Klein JP, Barrett AJ, et al. Severity of chronic graft-versus-host disease: association with treatment-related mortality and relapse. Blood 2002;100:406-14. [Crossref] [PubMed]
- Thomas ED, Storb R, Clift RA, et al. Bone-marrow transplantation (second of two parts). N Engl J Med 1975;292:895-902. [Crossref] [PubMed]
- Filipovich AH, Weisdorf D, Pavletic S, et al. National Institutes of Health consensus development project on criteria for clinical trials in chronic graft-versus-host disease: I. Diagnosis and staging working group report. Biol Blood Marrow Transplant 2005;11:945-56. [Crossref] [PubMed]
- Casto AM, Adler AL, Makhsous N, et al. Prospective, Real-time Metagenomic Sequencing During Norovirus Outbreak Reveals Discrete Transmission Clusters. Clin Infect Dis 2019;69:941-8. [Crossref] [PubMed]


