
Analysis of novel enzalutamide-resistant cells: upregulation of testis-specific Y-encoded protein gene promotes the expression of androgen receptor splicing variant 7
Introduction
Androgen depletion therapy (ADT) is performed for the treatment of prostate cancer (PCa). However, most patients with PCa will develop castration-resistant prostate cancer (CRPC) regardless of the treatment with ADT. Reactivation of androgen receptor (AR) and tumor recurrence occur in the presence of low androgen levels in patients with CRPC. Therefore, it is important to suppress the AR signal in CRPC. Based on these considerations, second-generation AR-targeted drugs, including abiraterone, a CYP17A1 inhibitor, and enzalutamide, a potent AR antagonist, were approved for the treatment of metastatic CRPC (mCRPC). Although these drugs have been shown to achieve prolongation of overall survival in patients with mCRPC before or after chemo-therapy (1-4), most patients with mCRPC develop resistance to abiraterone and enzalutamide. Various mechanisms of resistance to enzalutamide, including amplification of AR, overexpression of AR, AR splicing variants (AR-vs, such as AR-v7 and ARv567es), AR point mutations (F876L mutant), activation of AR by alternative pathways, and bypass pathway of AR signal, such as activation of glucocorticoid receptor (GR) (5,6), have been suggested in preclinical models. Some analyses of biomarkers of circulating tumor cells and cell-free tumor DNA in clinical studies have suggested that amplification of AR, expression of AR-v7, and AR mutation could be connected with resistance to enzalutamide (7-9). In terms of analyses in preclinical and clinical studies, aberrant activation of AR seems to play an important role in the resistance to enzalutamide, but the mechanisms through which it causes AR aberrations have not been clarified. Therefore, to develop new therapeutic strategies and facilitate drug discovery, it is important to explore the mechanisms of resistance to enzalutamide.
In the current study, we aimed to elucidate the mechanisms underlying resistance to enzalutamide in mCRPC. For this purpose, we established novel enzalutamide-resistant cells, namely SAS MDV No. 3–14 cells derived from human lymph node carcinoma of the prostate (LNCaP) cells. Then, we characterized the AR status and evaluated the gene expression profile in SAS MDV No. 3–14 cells. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-1463).
Methods
Reagents
Enzalutamide was synthesized in Taiho Pharmaceutical Co., Ltd. (Tokyo, Japan). Dihydrotestosterone (DHT) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Antibodies against AR (D6F11), AR-v7 (RM7), GAPDH, and testis-specific Y-encoded protein (TSPY; SF-7) were purchased from Cell Signaling Technology (Danvers, MA, USA), RevMAb Biosciences (San Francisco, CA, USA), Trevigen (Gaithersburg, MD, USA), and Santa Cruz Biotechnologies (Dallas, TX, USA), respectively. Fetal bovine serum (FBS) and charcoal-treated FBS (cFBS) were purchased from Thermo Fisher Scientific (Gibco, Waltham, MA, USA) and GE Healthcare (HyClone, Chicago, IL, USA), respectively.
Animals
Male BALB/c-nu mice (nude mice) for generating enzalutamide-resistant cells, were purchased from Charles River Laboratories (Wilmington, MA, USA). Male C.B-17/Icr-scid/scidJcl mice (SCID mice) for evaluating the effect of enzalutamide on LNCaP or enzalutamide-resistant tumor xenograft models in vivo, were purchased from Clea (Tokyo, Japan). Animals were housed under constant conditions of temperature (20–26 °C) with 12-hour lighting time (from 8:00 am to 8:00 pm) and provided sterilized food and water. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committees at Taiho Pharmaceutical Co., Ltd. (No. AE-2014-397 and AE-2016-179) and carried out according to the guidelines for animal experiments of Taiho Pharmaceutical Co., Ltd.
Cell culture
The PCa LNCaP cells were kindly provided by the Institute of Microbial Chemistry (Tokyo, Japan). Cells were maintained in RPMI1640 medium containing 10% FBS. For tumor implantation, LNCaP cells were treated with RPMI1640 medium containing 10% FBS and 5 ng/mL interleukin-6 (IL-6), as it has been reported that IL-6 increases the tumorigenic potential in vivo, and the xenograft mice model showed resistance to surgical castration (10). The SAS MDV No. 3–14 enzalutamide-resistant cells were maintained in RPMI1640 (phenol red free) medium with 5% FBS and 10 µM enzalutamide. Accordingly, VCaP, and 22Rv1 cells were obtained from the American Type Culture Collection and the European Collection of Authenticated Cell Cultures, respectively. Normal prostate stromal cells (PrSC) were purchased from LONZA (Tokyo, Japan).
Generation of enzalutamide-resistant cells
Enzalutamide-resistant cells were established in accordance with a previous study (11). Accordingly, LNCaP cells were subcutaneously inoculated in intact nude mice. Mice bearing tumors were surgically castrated under isoflurane anesthesia and administered 25 mg/kg enzalutamide for over 3 months. When the tumor volume reached about 2,000 mm3, tumors were excised, and viable tissue was transplanted to pre-castrated male nude mice. Second-generation transplanted xenografts were used for generating acquired enzalutamide-resistant cells. A relapsed tumor under continuous administration of enzalutamide was harvested by needle biopsy and cultured in vitro in the presence of 10 µM enzalutamide over the next 4 months, and thus SAS MDV No. 3–14 cells were established. In general, enzalutamide is administered at a dose of 10 mg/kg for evaluating its anti-tumor effect in preclinical studies (12,13). To generate enzalutamide resistant LNCaP xenografts, we adopted the dose of 25 mg/kg enzalutamide in the current study.
Cell growth assay
Cells were treated with indicated compounds at various concentrations in the presence or absence of DHT for 3 or 5 days. Cell proliferation was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA).
Reverse transcription and quantitative PCR analysis
Total RNA was extracted from cultured cells and purified using the RNeasy Mini Kit (QIAGEN, Venlo, The Netherlands). Reverse transcription PCR was performed with purified total RNA using the SuperScript VILO cDNA Synthesis Kit (Life Technologies, CA, USA), and cDNA was synthesized. Real-time quantitative reverse transcription PCR (qRT-PCR) was performed using the Applied Biosystems 7900HT Fast Real-Time PCR System and TaqMan Gene Expression Assays (Applied Biosystems, Waltham, MA, USA) for AR (Hs00171172_m1), AR-v7 (Hs04260217_m1), glyceraldehyde 3-phosphate dehydrogenase (GAPDH; Hs99999905_m1), kallikrein-3 (KLK3; Hs02576345_m1), FKBP prolyl isomerase 5 (FKBP5; Hs01561006_m1), transmembrane serine protease 2 (TMPRSS2; Hs01120965_m1), ubiquitin-conjugating enzyme E2 C (UBE2C; Hs00964100_g1), cyclin-dependent kinase 1 (CDK1; Hs00938777_m1), cell division cycle 20 (CDC20; Hs00426680_mH), and TSPY (Hs00413986_m1). Each mRNA expression level was normalized to the expression of GAPDH.
Immunoblotting analysis
Cultured cells were harvested and lysed in ice-cold M-PER Mammalian Protein Extraction Reagent (Thermo Fisher Scientific) containing a protease inhibitor cocktail (Sigma-Aldrich). Quantification of the protein content of cell lysates was carried out using a DC Protein Assay kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s instructions. An equal amount of protein (5 µg) was separated by SDS-PAGE electrophoresis and transferred to poly vinylidene difluoride (PVDF) membranes. Membranes were incubated in Blocking One (Nacalai Tesque, Kyoto, Japan) and probed with primary antibodies. After incubation with primary antibodies, membranes were incubated with an appropriate horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology). Chemiluminescence was detected using the ImageQuant LAS-4010 or Amersham Imager 600 systems (GE Healthcare).
RNA interference
The SAS MDV No. 3–14 cells were transfected with siRNAs targeting either AR-FL (5'-UCAAGGAACUCGAUCGUAU-3'), AR-v7 (5'-GUAGUUGUGAGUAUCAUGA-3'), AR-FL + AR-vs (ON-TARGETplus SMARTpool siRNA; Dharmacon, Lafayette, CO, USA), or TSPY (ON-TARGETplus SMARTpool siRNA; Dharmacon), including negative control RNA (ON-TARGETplus Non-targeting Control; Dharmacon) using an RNAi max reagent (Thermo Fisher Scientific) in the presence or absence of DHT.
Evaluation of the effect of enzalutamide on tumor growth in LNCaP or SAS MDV No. 3–14 xenograft models
Mice were surgically castrated under isoflurane anesthesia. LNCaP or SAS MDV No. 3–14 cells were subcutaneously inoculated in pre-castrated SCID mice. The mice were randomized into two groups and administered with vehicle or enzalutamide. Vehicle or 10 mg/kg of enzalutamide were orally administered for 21 (LNCaP) or 28 (SAS MDV No. 3–14) continuous days. Tumor volume was measured twice weekly.
Microarray analysis for gene expression profiling
Both LNCaP and SAS MDV No. 3–14 cells were treated with vehicle (DMSO) or DHT (1 nM) for 1 day. Then, total RNA was extracted from cultured cells and purified. Microarray analysis was performed using the Human Genome U133 Plus 2.0 Array (Thermo Fisher Scientific). Data were normalized using the robust multichip average. Differentially expressed genes in SAS MDV No. 3–14 cells treated with DMSO compared with those in LNCaP cells were extracted in a first step by filtering the expression ratio between LNCaP and SAS MDV No. 3–14 cells (absolute 2log ratio >2 difference). Next, DHT-regulated genes were excluded from the first-step-filtered genes. Obtained transcripts were used for Ingenuity Pathway Analysis® (IPA, QIAGEN), to explore the upstream genes of AR, already reported to exhibit a direct or indirect association with AR.
Statistical analysis
Statistical analysis was carried out using a Student’s t-test or Dunnett’s test. A P value <0.05 was considered to show statistical significance. All data analyses were performed using SAS version 9.4 and EXSUS version 10.0 (CAC Exicare Corporation, Tokyo, Japan).
Results
Generation of SAS MDV No. 3–14 enzalutamide-resistant cells
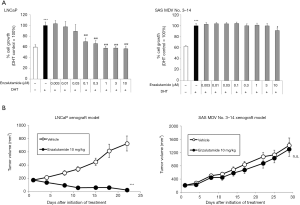
Nude mice bearing LNCaP cells were castrated and treated with enzalutamide over a period of 3 months. Progressive xenografts under treatment of enzalutamide were harvested and transplanted to pre-castrated nude mice. Treatment with enzalutamide showed tumor regression within a week; however, tumor reoccurrence was also observed. Relapsed xenograft tumors were harvested by biopsy with needle and cultured in vitro in the presence of 10 µM enzalutamide over a period of 4 months. The established cell line was named as SAS MDV No. 3–14 cells. To confirm the acquired resistance to enzalutamide in the generated SAS MDV No. 3–14 cell line, we compared the inhibitory effect of enzalutamide on the proliferation of both LNCaP and SAS MDV No. 3–14 cells. As shown in Figure 1A, treatment of LNCaP cells with enzalutamide (0.003–10 µM) suppressed cell growth in a dose-dependent manner. Contrastingly, treatment of SAS MDV No. 3–14 with enzalutamide had limited effect on cell growth. In addition, we evaluated the effect of enzalutamide in our in vivo models. Administration of enzalutamide resulted in tumor regression in our LNCaP xenograft model (Figure 1B), whereas enzalutamide did not exhibit an apparent effect on tumor growth in the SAS MDV No. 3–14 xenograft model.
Characterization of AR status and AR transactivation in SAS MDV No. 3–14 cells
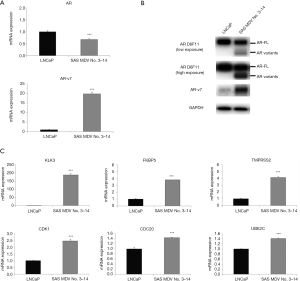
Since AR has been shown to play a pivotal role in the acquired resistance to enzalutamide, we aimed to characterize the AR status in SAS MDV No. 3–14 cells. We initially confirmed both the gene and protein expression levels of AR (Figure 2A,B). Accordingly, the mRNA expression of AR in SAS MDV No. 3–14 cells was comparable to that in LNCaP cells. However, the mRNA expression of AR-v7 in SAS MDV No. 3–14 cells was higher than that in LNCaP cells. These results were also confirmed in our protein expression analysis experiments. The expression of AR-FL protein in SAS MDV No. 3–14 was comparable to that in LNCaP cells, whereas the expression of AR-v7 protein was shown to be increased in SAS MDV No. 3–14 cells. To verify whether this differential AR status in SAS MDV No. 3–14 cells caused alterations in the transcriptional activity of AR, we next compared the expression of canonical AR (KLK3, FKBP5, and TMPRSS2) and AR-v7 (UBE2C, CDK1, and CDC20) target genes in SAS MDV No. 3–14 relative to that in LNCaP cells. The expression of KLK3, FKBP5, and TMPRSS2 in SAS MDV No. 3–14 cells was markedly higher than that in LNCaP cells. In addition, the expression of UBE2C, CDK1, and CDC20 in SAS MDV No. 3–14 cells was higher than that in LNCaP cells (Figure 2C).
The function of AR-FL and AR-v7 in cell growth and AR transactivation in SAS MDV No. 3–14 cells
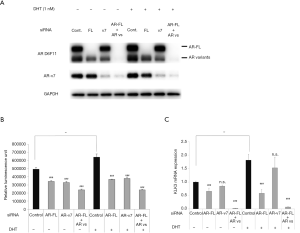
To evaluate the functional role of AR-FL and AR-v7 in SAS MDV No. 3–14 cells, we assessed the effects of knockdown by siRNA targeting AR-FL (targeting AR exon 7), AR-v7 (targeting AR cryptic exon 3), and AR-FL + AR-vs (targeting AR exon 1) on cell proliferation and the expression of KLK3, used as an index of the transcriptional activity of AR in the presence or absence of DHT. First, we validated the silencing of target sequences with each siRNA by western blot analysis (Figure 3A). We used an anti-AR n-terminal domain antibody (AR D6F11) and an anti-AR-v7 antibody to detect AR proteins. Western blot analysis demonstrated that AR-FL siRNA reduced the expression of AR-FL protein as detected by the AR D6F11 antibody. It also demonstrated that AR-v7 siRNA led to the reduced expression of AR-v7 protein detected by the AR-v7 antibody, as well as resulted in the moderately reduced expression of AR-vs as detected by the AR D6F11 antibody. Knockdown of AR-FL + AR-vs was shown to reduce the expression of both the AR-FL and AR-vs protein as detected by the AR D6F11 antibody. We next evaluated the effect of each siRNA on cell proliferation (Figure 3B). Knockdown of AR-FL and AR-v7 led to reduced cell proliferation in the presence and absence of DHT. Further, AR siRNA targeting AR-FL + AR-vs was shown to suppress cell proliferation more potently than each of the knockdowns of AR-FL or AR-v7 regardless of the presence or absence of DHT. Furthermore, we evaluated the effect of each siRNA on the expression of KLK3, as an index of the transactivation of AR. Knockdown of AR-FL resulted in the reduced expression of KLK3 in the presence and absence of DHT (Figure 3C), whereas AR-v7 siRNA only moderately suppressed the expression of KLK3. In contrast, AR siRNA targeting AR-FL + AR-v7 was shown to markedly reduce the expression of KLK3 in the presence and absence of DHT.
Microarray analysis for a comprehensive evaluation of altered gene expression in SAS MDV No. 3–14 compared with LNCaP cells
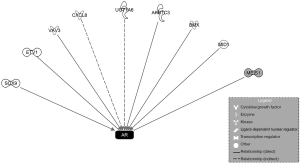
To comprehensively explore the differential gene expression associated with the acquired resistance to enzalutamide in SAS MDV No. 3–14 cells, we performed microarray analysis. First, we selected genes whose expression in SAS MDV No. 3–14 cells was different from that in LNCaP cells. Selected genes were differentially expressed if the absolute 2log ratio was greater than 2. According to the criteria, we identified 566 differentially expressed transcripts in SAS MDV No. 3–14 compared with LNCaP cells. We next excluded from further analysis any first-step-identified androgen-dependent genes, whose expression was regulated by DHT treatment in both cells. Consecutively, we got 294 extracted genes representing a set of candidate genes that might be responsible for the acquired resistance to enzalutamide in SAS MDV No. 3–14 cells. The top 30 differentially upregulated transcripts in SAS MDV No. 3–14 compared with LNCaP cells are shown in Table 1. There were several genes [transmembrane protein with EGF like and two follistatin like domains 2 (TMEFF2), HIV-1 Tat interactive protein 2 (HTATIP2), and UDP glucuronosyltransferase family 1 member A (UGT1A)] whose expression levels are altered in CRPC (14-16), included in the list. From the top 30 genes in our list, we chose to evaluate the function of TSPY in SAS MDV No. 3–14 cells, as it was one of the highest upregulated genes. It has been reported that TSPY could stimulate the transactivation of AR (17). However, the function of TSPY in the mechanisms underlying the acquired resistance to enzalutamide has not been elucidated. In addition, we performed IPA using these extracted genes to explore the differential gene expression of upstream components of AR. Accordingly, 9 genes [SRY-box transcription factor 9 (SOX9), ETS variant transcription factor 1 (ETV1), vav guanine nucleotide exchange factor 3 (VAV3), C-X-C motif chemokine ligand 8 (CXCL8), UDP-glucuronosyltransferase family 1 member 6 (UGT1A6), aldo-keto reductase family 1 member C3 (AKR1C3), BMX non-receptor tyrosine kinase (BMX), midline 1 (MID1), and meis homeobox 1 (MEIS1)] upstream of AR were identified as differentially expressed in SAS MDV No. 3–14 cells (Figure 4). The VAV3, SOX9 and AKR1C3 upregulated genes identified in SAS MDV No. 3–14 cells, have already been reported as genes responsible for the progression of CPRC or acquired resistance to enzalutamide (18-20). Therefore, we prioritized the analysis of TSPY in the current study.
Table 1
| 2log ratio | Gene symbol |
|---|---|
| 7.91 | TMEFF2 |
| 7.45 | WDR72 |
| 7.09 | GJA1 |
| 6.62 | WBP5 |
| 6.55 | KRTAP3-3 |
| 6.45 | KCNJ3 |
| 6.11 | HTATIP2 |
| 6.08 | KCNJ3 |
| 5.79 | TSPY1, TSPY10, TSPY2, TSPY3, TSPY4, TSPY8 |
| 5.75 | IL32 |
| 5.71 | ADGRG6 |
| 5.66 | UGT1A1, UGT1A10, UGT1A3, UGT1A4, UGT1A5, UGT1A6, UGT1A7, UGT1A8, UGT1A9 |
| 5.48 | LRRN1 |
| 5.46 | TENM2 |
| 5.40 | SCCPDH |
| 5.38 | UGT1A1, UGT1A10, UGT1A3, UGT1A4, UGT1A5, UGT1A6, UGT1A7, UGT1A8, UGT1A9 |
| 5.37 | TUSC1 |
| 5.31 | ST8SIA4 |
| 5.21 | UGT1A1, UGT1A10, UGT1A4, UGT1A6, UGT1A8, UGT1A9 |
| 5.05 | UGT1A1, UGT1A10, UGT1A4, UGT1A6, UGT1A8, UGT1A9 |
| 5.01 | SFN |
| 4.88 | CCDC169-SOHLH2, SOHLH2 |
| 4.85 | KCNJ3 |
| 4.70 | CCL8 |
| 4.60 | ZNF320 |
| 4.57 | CCL2 |
| 4.37 | NR2F1 |
| 4.34 | S1PR3 |
| 4.32 | ST8SIA4 |
| 4.28 | ETV1 |
TMEFF2, transmembrane protein with EGF like and two follistatin like domains 2; WDR72, WD repeat-containing protein 72; GJA1, gap junction alpha-1 protein; WBP5, WW domain binding protein 5; KRTAP3-3, keratin-associated protein 3-3; KCNJ3, potassium inwardly-rectifying channel, subfamily J, member 3; HTATIP2, HIV-1 Tat interactive protein 2; TSPY, testis-specific Y-encoded protein; IL32, Interleukin 32; ADGRG6, adhesion G-protein coupled receptor G6; UGT1A, glucuronosyltransferase family 1 member A1; LRRN1, leucine-rich repeat neuronal protein 1; TENM2, teneurin-2; SCCPDH, saccharopine dehydrogenase-like oxidoreductase; TUSC1, tumor suppressor candidate gene 1; ST8SIA4, CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase; SFN, 14-3-3 protein sigma; CCDC169-SOHLH2, CCDC169-SOHLH2 isoform 1; CCL8, C-C motif chemokine 8; ZNF320, Zinc finger protein 320; CCL2, C-C motif chemokine 2; NR2F1, COUP transcription factor 1; S1PR3, sphingosine 1-phosphate receptor 3; ETV1, ETS translocation variant 1.
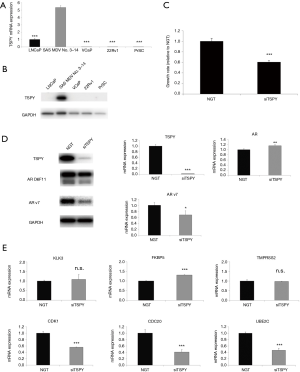
Evaluation of the function of TSPY in SAS MDV No. 3–14 cells
To validate the differential expression of the TSPY gene between SAS MDV No. 3–14 and LNCaP cells in our microarray analysis, we first confirmed the expression of both the TSPY mRNA and protein using qRT-PCR and western blot analysis in SAS MDV No. 3–14 cells, other PCa cell lines, and normal prostate cells. The expression of TSPY mRNA was higher in SAS MDV No. 3–14 than that in LNCaP cells, other PCa cells (VCaP, 22Rv1), and normal PrSC (Figure 5A). Likewise, the expression of TSPY protein was also higher in SAS MDV No. 3–14 cells than that of other cell lines (Figure 5B). To evaluate the role of TSPY in SAS MDV No. 3–14 cells, we assessed the effect of a knockdown of TSPY on cell growth, expression of AR, and expression of AR target genes, using a siRNA targeting the TSPY gene. Accordingly, TSPY siRNA inhibited the proliferation of SAS MDV No. 3–14 cells (Figure 5C). Western blot analysis demonstrated that knockdown of TSPY led to reduced expression of AR-v7 in SAS MDV No. 3–14 cells (Figure 5D). Contrarily, the knockdown of TSPY did not exhibit any apparent effect on AR-FL and AR variants, other than AR-v7. Similar results were also obtained from our qRT-PCR analysis of the mRNA expression (Figure 5D). Silencing of the expression of TSPY did not show any apparent effects on the canonical expression of AR target genes, including KLK3, FKBP5, and TMPRSS2 (Figure 5E). In contrast, knockdown of TSPY reduced the expression of AR-v7 target genes, such as CDK1, CDC20, and UBE2C (Figure 5E).
Discussion
Resistance to enzalutamide constitutes a clinical issue in the management and treatment of patients with CRPC. Several enzalutamide-resistant models that harbor AR aberrations, including amplification of AR, overexpression of AR, AR splicing variants, such as AR-v7, and AR mutations in the AR ligand-binding domain (LBD) (F876L) have been reported (21-23). However, the mechanisms of resistance have not been fully elucidated. To solve this problem, we established a new enzalutamide-resistant cell line, named SAS MDV No. 3–14 cells, and performed a gene expression profiling of these cells.
Our SAS MDV No. 3–14 cells were shown to express AR-FL, AR-v7, and other AR-vs (Figure 2A,B), and to harbor increased transcriptional activity of AR, including upregulation of canonical AR (KLK3, FKBP5, and TMPRSS2) and AR-v7 (CDK1, CDC20, and UBE2C) target genes compared with LNCaP cells. Concomitantly, experiments of knockdown of AR revealed that both AR-FL and AR-v7 could regulate the proliferation of SAS MDV No. 3–14 cells (Figure 3B), whereas AR-FL and AR-vs could predominantly regulate the transcriptional activity of AR (Figure 3C). It has been reported that 22Rv1 and JDCaP-hr cells express AR-FL and AR-v7, and knockdown of AR-v7 was shown to predominantly suppress cell growth and the transcriptional activity of AR in each cell line (22,24). Similar to these reported cells, the proliferation of SAS MDV No. 3–14 cells was partially regulated through AR-v7. In addition, it was demonstrated that not only AR-v7, but also AR-FL could play this pivotal role in the proliferation of SAS MDV No. 3–14 cells. Several previous studies demonstrated that a gain in the AR copy number, mutations on the AR LBD, especially F876L, and overexpression of GR could cause acquired resistance to enzalutamide (21,23,25). However, neither a gain in the AR copy number, mutations on AR LBD, except for T877A, or overexpression of GR were detected in our SAS MDV No. 3–14 cells (data not shown). These results indicated that SAS MDV No. 3–14 cells, whose proliferation mainly depends on both AR-FL and AR-v7, are unique in their acquired resistance to enzalutamide.
Previous studies adopted a global approach to identify adaptive changes in gene expression related to the acquisition of resistance to enzalutamide (20,26). We also performed microarray analysis to comprehensively explore the differences in gene expression between SAS MDV No. 3–14 and LNCaP cells. In our study, TSPY was one of the highest upregulated genes in SAS MDV No. 3–14 cells compared with LNCaP cells. In addition, TSPY has been reported as an activator of the transcription of AR (17). However, it remains uncertain whether TSPY might be associated with the acquired resistance to enzalutamide. Therefore, we prioritized the analysis of the function of TSPY in SAS MDV No. 3–14 cells using siRNA approaches. Notably, TSPY is known to be usually expressed during spermatogonia of fetal testis and adult testis (27). In contrast, TSPY also exerts significant influence on the oncogenic process of somatic cancer, including PCa. The expression level of TSPY could be correlated with Gleason grading in PCa specimens (28). It has been reported that TSPY accelerates cell proliferation, and G2/M transition through activation of cyclin-dependent kinase 1, suggesting the function of TSPY as a cell cycle regulator (29). In the current study, the knockdown of TSPY was shown to reduce the expression of AR-v7 in SAS MDV No. 3–14 cells, resulting in the downregulation of the expression of AR-v7 target genes and suppression of cell growth. As such, our study discovered a novel function of TSPY as a positive regulator of the expression of AR-v7 in SAS MDV No. 3–14 cells. In addition, our study also demonstrated that TSPY is one of the genes involved in the acquired resistance to enzalutamide. Various factors leading to the acquisition of expression of AR-v7 in PCa cells, such as p52 and Sam68, have also been reported (30,31). We confirmed that knockdown of TSPY did not have an apparent effect on the expression of either p52, or Sam68 proteins (data not shown). Further studies are needed to verify the mechanisms through which TSPY generates the expression of AR-v7 in SAS MDV No. 3–14 cells.
Among the top 30 differentially upregulated transcripts in both our microarray (Table 1) and IPA pathway analyses, we identified several genes potentially associated with the progression of CRPC or acquired resistance to enzalutamide, including TMEFF2, HTATIP2, UGT1A, VAV3, SOX9, and AKR1C3 (14-16,18-20), in SAS MDV No. 3–14 cells. These genes could contribute to the acquired resistant to enzalutamide and to the enhancement of the transcriptional activity of AR in SAS MDV No. 3–14 cells. It has also been reported that non-AR mediated mechanisms, such as bypassing of AR signaling, lineage switching, and tumor microenvironment changes could be associated with the resistance to enzalutamide (5). However, the contribution of these non-AR mediated resistance mechanisms to SAS MDV No. 3–14 cells has not been fully understood. Further experiments would be required to fully characterize SAS MDV No. 3–14 cells and elucidate the mechanisms underlying the acquisition of resistance to enzalutamide.
Conclusions
In our study, we established SAS MDV No. 3–14 cells as a novel enzalutamide-resistant cell line derived from human PCa LNCaP cells. Our results demonstrated that SAS MDV No. 3–14 cells express AR-v7, and both AR-FL and AR-v7 simultaneously function as drivers of cell growth in these cells. In addition, we discovered that TSPY positively regulates the expression of AR-v7 in SAS MDV No. 3–14 cells. Therefore, SAS MDV No. 3–14 cells could provide valuable information toward clarifying the mechanisms underlying the resistance to enzalutamide and toward analyzing the mechanism of acquired expression of AR-v7, as well as facilitate drug discovery and development of novel therapies for CRPC.
Acknowledgments
The authors thank Dr. Tsutomu Kobayashi and Ms. Kayo Narukawa for their insightful discussions.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-1463
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1463
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form. (Available at http://dx.doi.org/10.21037/tcr-20-1463). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committees at Taiho Pharmaceutical Co., Ltd. (No. AE-2014-397 and AE-2016-179) and carried out according to the guidelines for animal experiments of Taiho Pharmaceutical Co., Ltd.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012;367:1187-97. [Crossref] [PubMed]
- Beer TM, Armstrong AJ, Rathkopf DE, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 2014;371:424-33. [Crossref] [PubMed]
- de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011;364:1995-2005. [Crossref] [PubMed]
- Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013;368:138-48. [Crossref] [PubMed]
- Vander Ark A, Cao J, Li X. Mechanisms and Approaches for Overcoming Enzalutamide Resistance in Prostate Cancer. Front Oncol 2018;8:180. [Crossref] [PubMed]
- Galletti G, Leach BI, Lam L, et al. Mechanisms of resistance to systemic therapy in metastatic castration-resistant prostate cancer. Cancer Treat Rev 2017;57:16-27. [Crossref] [PubMed]
- Salvi S, Casadio V, Conteduca V, et al. Circulating AR copy number and outcome to enzalutamide in docetaxel-treated metastatic castration-resistant prostate cancer. Oncotarget 2016;7:37839-45. [Crossref] [PubMed]
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028-38. [Crossref] [PubMed]
- Wyatt AW, Azad AA, Volik SV, et al. Genomic Alterations in Cell-Free DNA and Enzalutamide Resistance in Castration-Resistant Prostate Cancer. JAMA Oncol 2016;2:1598-606. [Crossref] [PubMed]
- Lee SO, Lou W, Hou M, et al. Interleukin-6 promotes androgen-independent growth in LNCaP human prostate cancer cells. Clin Cancer Res 2003;9:370-6. [PubMed]
- Kuruma H, Matsumoto H, Shiota M, et al. A novel antiandrogen, Compound 30, suppresses castration-resistant and MDV3100-resistant prostate cancer growth in vitro and in vivo. Mol Cancer Ther 2013;12:567-76. [Crossref] [PubMed]
- Tran C, Ouk S, Clegg NJ, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 2009;324:787-90. [Crossref] [PubMed]
- Clegg NJ, Wongvipat J, Joseph JD, et al. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res 2012;72:1494-503. [Crossref] [PubMed]
- Overcash RF, Chappell VA, Green T, et al. Androgen signaling promotes translation of TMEFF2 in prostate cancer cells via phosphorylation of the α subunit of the translation initiation factor 2. PLoS One 2013;8:e55257. [Crossref] [PubMed]
- Singh AP, Bafna S, Chaudhary K, et al. Genome-wide expression profiling reveals transcriptomic variation and perturbed gene networks in androgen-dependent and androgen-independent prostate cancer cells. Cancer Lett 2008;259:28-38. [Crossref] [PubMed]
- Williamson SC, Mitter R, Hepburn AC, et al. Characterisations of human prostate stem cells reveal deficiency in class I UGT enzymes as a novel mechanism for castration-resistant prostate cancer. Br J Cancer 2013;109:950-6. [Crossref] [PubMed]
- Li Y, Zhang DJ, Qiu Y, et al. The Y-located proto-oncogene TSPY exacerbates and its X-homologue TSPX inhibits transactivation functions of androgen receptor and its constitutively active variants. Hum Mol Genet 2017;26:901-12. [Crossref] [PubMed]
- Peacock SO, Fahrenholtz CD, Burnstein KL. Vav3 enhances androgen receptor splice variant activity and is critical for castration-resistant prostate cancer growth and survival. Mol Endocrinol 2012;26:1967-79. [Crossref] [PubMed]
- Khurana N, Sikka SC. Interplay Between SOX9, Wnt/β-Catenin and Androgen Receptor Signaling in Castration-Resistant Prostate Cancer. Int J Mol Sci 2019;20:E2066. [Crossref] [PubMed]
- Liu C, Yang JC, Armstrong CM, et al. AKR1C3 Promotes AR-V7 Protein Stabilization and Confers Resistance to AR-Targeted Therapies in Advanced Prostate Cancer. Mol Cancer Ther 2019;18:1875-86. [Crossref] [PubMed]
- Hoefer J, Akbor M, Handle F, et al. Critical role of androgen receptor level in prostate cancer cell resistance to new generation antiandrogen enzalutamide. Oncotarget 2016;7:59781-94. [Crossref] [PubMed]
- Nakata D, Nakayama K, Masaki T, et al. Growth Inhibition by Testosterone in an Androgen Receptor Splice Variant-Driven Prostate Cancer Model. Prostate 2016;76:1536-45. [Crossref] [PubMed]
- Balbas MD, Evans MJ, Hosfield DJ, et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. Elife 2013;2:e00499. [Crossref] [PubMed]
- Li Y, Chan SC, Brand LJ, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013;73:483-9. [Crossref] [PubMed]
- Arora VK, Schenkein E, Murali R, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013;155:1309-22. [Crossref] [PubMed]
- Kregel S, Chen JL, Tom W, et al. Acquired resistance to the second-generation androgen receptor antagonist enzalutamide in castration-resistant prostate cancer. Oncotarget 2016;7:26259-74. [Crossref] [PubMed]
- Lau YC, Li Y, Kido T. Battle of the sexes: contrasting roles of testis-specific protein Y-encoded (TSPY) and TSPX in human oncogenesis. Asian J Androl 2019;21:260-9. [Crossref] [PubMed]
- Kido T, Hatakeyama S, Ohyama C, et al. Expression of the Y-Encoded TSPY is Associated with Progression of Prostate Cancer. Genes (Basel) 2010;1:283-93. [Crossref] [PubMed]
- Oram SW, Liu XX, Lee TL, et al. TSPY potentiates cell proliferation and tumorigenesis by promoting cell cycle progression in HeLa and NIH3T3 cells. BMC Cancer 2006;6:154. [Crossref] [PubMed]
- Nadiminty N, Tummala R, Liu C, et al. NF-κB2/p52 induces resistance to enzalutamide in prostate cancer: role of androgen receptor and its variants. Mol Cancer Ther 2013;12:1629-37. [Crossref] [PubMed]
- Stockley J, Markert E, Zhou Y, et al. The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci Rep 2015;5:13426. [Crossref] [PubMed]


