The clinical significance of RET gene fusion among Chinese patients with lung cancer
Introduction
Lung cancer is the leading cause of cancer-associated death worldwide, consisting of over 50 histomorphology subtypes. Non-small cell lung cancer (NSCLC) is the most common subtype in lung cancer and adenocarcinoma (ADC), along with squamous cell carcinoma (SqCC) accounts for the major histological subtypes. Most of the patients are diagnosed with distant metastasis which made the total resection difficult to achieve. The combination of histology and molecular biomarkers analysis refined the current classification of lung cancer. Epidermal growth factor receptor (EGFR) mutations were found in approximately 43–89% of patients with NSCLC. Human epidermal growth factor receptor 2 (HER2) exon 20 mutations, analogous to the exon 20 mutations in EGFR, was identified as an actionable target in NSCLC development. Multiple novel driver gene mutations including hepatocyte growth factor receptor (MET) exon 14 mutations, proto-oncogene tyrosine-protein kinase receptor Ret (RET), neurotrophic tyrosine kinase (NTRK) fusion, KRAS mutations, and neurofibromin 1 (NF1) loss were identified and most of them mutated exclusively. Serine/threonine-protein kinase b-raf (BRAF), was a newly FDA-approved therapy target (1-3).
The proto-oncogene RET was located on chromosomal 10q11.2 and encoded a single-pass transmembrane receptor tyrosine receptor. Previous results highlighted the critical role of the RET gene in the development and maintenance of several tissue and cell types (4). RET was initially found in NIH-3T3 cells that were transfected with lymphoma DNA and then detected in papillary thyroid cancers. About 1–2% of the patients with lung cancer were positive for the mutation and it occurred more frequently in the non-smoker patients with ADC (5). Kinesin family member 5b (KIF5B) and coiled-coil domain containing 6 (CCDC6) were the common upstream fusion partners of RET fusions in NSCLC. The chromosomal rearrangement could contribute to the constitutive expression of the RET fusion protein and activate the survival associated pathways (6).
Targeted therapy has become part of the routine management of patients with positive driver gene mutations recently. Targeted drugs including tyrosine kinase inhibitors (TKI) of EGFR mutation, inhibitors of anaplastic lymphoma kinase (ALK) rearrangement, anti-angiogenesis agents, antibodies against vascular endothelial growth factor (VEGF) were approved for NSCLC treatment (7,8). The targetable fusion proteins in NSCLC were ALK, ROS1, NTRK, and RET (9). Several multi-targets RET inhibitors were demonstrated the anti-tumor effect in cell lines and xenograft with RET fusion. Clinical trials have proved the efficacy of cabozantinib (10) and vandetanib (11,12) on NSCLC patients with RET fusions. Results of a retrospective study from a global multicenter registry in NSCLC patients with RET fusions suggested that the anti-tumor activity of RET inhibitors were limited compared to targeted therapy in EGFR mutant and ALK/ROS1-rearranged patients with lung cancer (5). A retrospective study enrolled 6,125 samples summarized the clinical and molecular features of RET fusions in Chinese patients with NSCLC. However, the therapeutic efficacy of RET inhibitors in these patients remained unknown (13). Therefore, we analyzed 39 NSCLC samples with RET fusions using next-generation sequencing and collected the anonymized data. We tried to figure out the molecular features of RET fusion in Chinese patients with NSCLC. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-754).
Methods
Patient information
A total of 39 samples with RET fusions were collected from Hunan Provincial Tumor Hospital, Tianjin Cancer Institute, and Chinese Academy of Medical Science Tumor Hospital from 2016 to 2018. The cell origin was identified using histological analysis according to the 4th World Health Organization classification (14). Patients characteristics including gender, age, or clinical stage were collected. The cancer stages were evaluated according to the 8th TNM staging system. the overall survival (OS) data were available in 21 samples and progression-free data were available in 14 samples. The treatment strategies of each patient were obtained from the medical records. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Research Ethics Committee of Cancer Hospital Chinese Academy of Medical Sciences (No. 18-118/1696) and informed consent was taken from all the patients.
Targeted DNA sequencing
DNA of formalin-fixed, paraffin-embedded (FFPE) samples were extracted using QIAamp DNA FFPE tissue kit (QIAGEN, Valencia, CA) and the DNA concentration was measured by Qubit dsDNA assay. The gDNA quality was assessed to make sure that A260/A280 is within the range of 1.8 to 2.0. For patients with available DNA, targeted DNA sequencing was performed. DNA was profiled using the Lung Plasma panel (Burning Rock Biotech) covering 168 cancer-associated genes. The concentration of the DNA samples was measured with the Qubit dsDNA assay to make sure that genomic DNA was greater than 40 ng. Fragments of 200 to 400-bp sizes were selected with beads (Agencourt AMPure XP kit; Beckman-Coulter, Brea, CA), followed by hybridization with the capture probes baits, hybrid selection with magnetic beads, and PCR amplification. A bioanalyzer high-sensitivity DNA assay was then used to evaluate the quality and size range. Available indexed samples were then sequenced on a Miseq (Illumina, San Diego, CA) with pair-end reads.
Sequencing data analysis
Sequencing data were mapped to the human genome (hg19) using BWA aligner 0.7.10 (15). PCR duplicate reads were removed before base substitution detection. Local alignment optimization and variant calling was performed using GATK v3.2-2 (16). DNA translocation analysis was performed using both Tophat2 and Factera 1.4.3 (17,18). Insert size distribution and library complexity of each sample were computed to assess the level of DNA degradation. Different mutation calling thresholds were applied on samples with different DNA quality to avoid false-positive mutation calls due to DNA damage. SNV and indels identification were annotated using the dbNSFP(v30a), COSMIC (v69), and dbSNP (snp138) database. Variants with a global minor allele frequency greater than 1.0% in 1000Genome Project (Phase3, http://www.1000genomes.org/data) were considered as common SNPs and removed. Integrative Genomics Viewer (Broad Institute, USA) was used to visualize variants aligned against the reference genome to confirm the accuracy of the variant calls by checking for possible strand biases and sequencing errors. Gene-level copy number variation (CNV) was assessed using a statistic after normalizing read depth at each region by total read number and region size and correcting GC-bias using a LOESS algorithm (19).
Statistical analysis
GraphPad Prism 5 was used for graphs and statistical analysis. The data of proportion variables were recorded by frequency or percentage. PFS was defined as the time between RET inhibitor treatment initiation to disease progression evaluated by doctors or death from any cause. OS was the time from disease diagnosis to death from any cause. For those without progression or death event by the end of the study, survival end points were censored at the date of last follow-up. The survival differences were performed using the log-rank test and Kaplan-Meier method was used to estimate the survival curves.
Results
Patients characteristics
To better understand the therapeutic efficacy of RET inhibitors in lung ADC patients harboring RET mutations, we collected 39 samples and conducted five-year survival follow-up. Of the 39 subjects included, 19 (48.72%) samples were male and 20 (51.28%) of the patients were female. 76.92% of all samples was under 65 years, ranging from 35 to 82 years with the median age of 59 years old. Most of the samples (36 of 39 patients; 92.31%) were diagnosed with lung adenocarcinoma (LUAD) and one was diagnosed with lung squamous cell carcinoma (LUSC). The tumor stages in our cohort ranged from I to IV and over half of the samples were staged IV. Eight (20.51%) samples were stage III, 3 (7.69%) were stage II and 3 (7.69%) were stage I. The treatment methods of each patient were also recorded. Thirteen patients with RET fusions received RET inhibitors. Six (46.15%) of them were females and 7 (53.84%) were males. Most (12 of 13 patients; 92.31%) of them had advanced stage III and IV disease (Table 1).
Table 1
| Patient characteristics | Total | RET inhibitors | KIF5B-RET | |||
|---|---|---|---|---|---|---|
| Treated (n=13) | Non-treated (n=26) | Positive (n=23) | Negative (n=16) | |||
| Gender, n (%) | ||||||
| Male | 19 (48.72) | 6 (46.15) | 13 (50.00) | 10 (43.48) | 9 (56.25) | |
| Female | 20 (51.28) | 7 (53.84) | 13 (50.00) | 13 (56.52) | 7 (43.75) | |
| Age (years), n (%) | ||||||
| <65 | 30 (76.92) | 10 (76.92) | 20 (76.92) | 16 (69.57) | 14 (87.50) | |
| ≥65 | 9 (23.08) | 3 (23.08) | 6 (23.08) | 7 (30.43) | 2 (12.50) | |
| Histological types, n (%) | ||||||
| Lung adenocarcinoma (LUAD) | 36 (92.31) | 11 (84.61) | 25 (96.15) | 21 (91.30) | 15 (93.75) | |
| Other lung cancers | 3 (7.69) | 2 (15.38) | 1 (3.84) | 2 (8.70) | 1 (6.25) | |
| Tumor stages, n (%) | ||||||
| Ia | 2 (5.13) | 0 | 2 (7.69) | 1 (4.35) | 1 (6.25) | |
| IIb | 1 (2.56) | 0 | 1 (3.85) | 1 (4.35) | 0 | |
| IIa | 1 (2.56) | 1 (7.69) | 0 | 1 (4.35) | 0 | |
| IIb | 2 (5.13) | 1 (7.69) | 1 (3.85) | 1 (4.35) | 1 (6.25) | |
| IIIa | 4 (10.25) | 3 (23.08) | 1 (3.85) | 1 (4.35) | 3 (18.75) | |
| IIIb | 3 (7.69) | 0 | 3 (11.54) | 3 (13.04) | 0 | |
| IV | 25 (64.10) | 9 (69.23) | 16 (61.54) | 15 (65.22) | 10 (62.50) | |
RET fusion overview
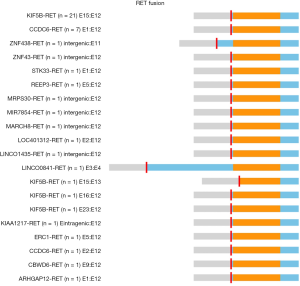
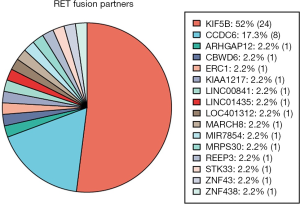
A total of 46 RET fusions in 39 patients were identified and 24 (52%) patients harbored KIF5B-RET fusion. In this cohort, the most common variant (21 of 24 variants, 87.5%) of KIF5B-RET fusion was KIF5B exon 15 fused to RET exon 12 (K15-R12) variant. The other three were K15-R13, K16-R12, and K23-R12 respectively (Figure 1). CCDC6 was the second common partner of RET fusion as 8 (20.51%) patients were positive for this variant. Several new and rare RET fusions occurred in one patient including that RET fused with ZNF438, ZNF43, STK33, REEP3, MRPS30, MIR7854, MARCH8, LOC401312, LINC01435, LINC00841, KIAA1217, ERC1, CBWD6 and ARHGAP12 (Figure 2) (Table S1). Of eight samples who harbored two RET fusions, five were positive for KIF5B-RET fusion and three were positive for CCDC6-RET fusions.
Concurrent genomic alterations in patients with positive RET-fusion
A total of 8 samples in this cohort were positive with at least two variants in this panel. TP53 was the most common concurrent gene mutation, 6 (15%) samples in this cohort harbored TP53 mutation along with the RET fusions. Three samples harbored three variants which all belonged to the RET fusion and TP53 concurrent mutation group. And the third variant occurred in GRIN2A, NAV3, and SOX9, respectively. Regarding the RET fusions that occurred with TP53 mutation, concurrent mutations in ERC1, CCDC6, and KIF5B were all observed in our cohort. Besides, we observed the concurrent mutations in ERBB2 (1/39) and NTRK1 (1/39). ERBB2 mutation and KIF5B-RET fusion were only noted in a patient with pulmonary sarcomatoid carcinoma (PSC). NTRK1 mutation was found to cooccur with CCDC6-RET and LINC01435-RET fusions. Three samples accounting for 8% of all the cases harbored EGFR mutation and one sample was positive with exon 19 EGFR mutation (Figure 3).

Clinical outcomes of patients receiving RET inhibitors treatment
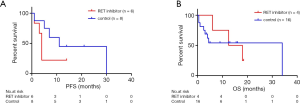
Cabozantinib was administered in nine patients, among whom one was treated with erlotinib and cabozantinib and three patients were received Anlotinib or Crizotinib. Of those who were available for the response to Cabozantinib, 2 patients (20%; 0–45) with respective KIF5B-RET and ERC1-RET fusions experienced disease progression, 2 (20%; 0–45) patients with KIF5B-RET fusions achieved stable disease (SD) and 1 (10%; 0–29) patient was not evaluable. However, no patient achieved partial or complete response (CR). Twenty-six patients with RET fusions were not administered RET inhibitors. Among these patients, 3 patients achieved partial response (11.54%; 0–24) including 2 with KIF5B-RET fusion and 1 with CCDC6-RET fusion. Stable disease was noted in 4 patients (15.38%; 2–29), 2 subjects harbored KIF5B-RET fusions, 1 had two RET fusion partners including ARHGAP12 and KIF5B and the other was positive for CCDC6-RET fusion. The PFS and OS data were available for 6 patients and 5 patients respectively in the group who received Cabozantinib. The median PFS and OS were 4 months (95% CI, 3.2–4.8) and 25 months (95% CI, 1.48–48.52). With regard to the patients who did not receive RET inhibitors, the PFS and OS data were available for 8 patients and 16 patients. The median PFS was 11 months (95% CI, 1.16–20.84) (Table 2). Survival comparison between patients who received and not received RET inhibitors indicated that there was no significant difference between the two groups in terms of PFS (Figure 4A) and OS (Figure 4B).
Table 2
| Drug | Complete response (%; 95% CI) | Partial response (%; 95% CI) | Stable disease (%; 95% CI) | Disease progression (%; 95% CI) | Not evaluable (%; 95% CI) | Missing data (%; 95% CI) | Median PFS (95% CI) | Median OS (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Cabozantinib (n=10) | 0 | 0 | 2 (20%; 0–45) | 2 (20%; 0–45) | 1 (10%; 0–29) | 5 (50%; 19–81) | 4 (3.2–4.8) | 25 (1.48–48.52) |
| Anlotinib (n=2) | 0 | 0 | 0 | 0 | 0 | 2 | ||
| Crizotinib (n=1) | 0 | 0 | 0 | 0 | 0 | 1 | ||
| RET inhibitor not treated (n=26) | 0 | 3 (11.54%; 0–24) | 4 (15.38%; 2–29) | 0 | 3 (11.54%; 0–24) | 16 (61.54%; 43–80) | 11 (1.16–20.84) | Not reached |
Discussion
Several in vitro and in vivo studies have identified the potential significance of RET as a promising targetable driver gene in NSCLC. However, the efficacy of RET inhibitors in Chinese patients remained unclear (9). A previous large-scale study in lung cancer patients suggested that RET fusions occurred in about 1–2% among East Asian patients (20). A similar mutation rate was observed in a recent study including 6,125 Chinese lung cancer patients in which 1.4% of the samples were positive with RET fusions (13). Taking the rare mutation rate of RET fusions into consideration, it was hard to generate meaningful clinical data in a clinical study with small sample size. We collected 39 samples with a least one RET fusion detected using NGS. In our study, most of the samples were diagnosed with LUAD and the median age of the patients at diagnosis was 59 years. The results were consistent with most studies in which RET fusion prone to occur in patients with LUAD at younger age. Another meta-analysis including 84 patients with RET fusions showed a higher mutation frequency of RET fusion in younger, non-smoking female patients especially in Asian area (21). However, there was some discrepancy in the association between gender and RET fusion. A study in the Japanese cohort showed no difference in the frequency of RET fusion between male and female patients (22). Another study in the European cohort reported a higher frequency of the male patients than the female patients (23). Our study involved 19 (48.72%) female patients and 20 (51.28%) male patients. The discrepancy may come from the difference in ethnicity, environment and sample size. Most of the samples we collected were diagnosed with advanced lung cancer as 84.61% of the patients were stage IIIb and IV, which highlighted the potential role of RET mutation in advanced NSCLC treatment.
In lung tissues, KIF5B was dominantly expressed and functioned to activate the ALK/RET tyrosine kinase and downstream oncogenic effector. In NSCLC, KIF5B was the fusion partner of ALK, protein tyrosine kinase (PTK), and RET (24). KIF5B-RET fusion accounted for 2% of patients with NSCLC. Takashi Kohno et. al firstly identified KIF5B-RET fusion in patients with lung cancer from Japan and the United States using whole-transcriptome sequencing. They found an increase of RET expression in cells with KIF5B-RET fusion (25). The most common KIF5B-RET variant was K15-R12 which accounted for 87.5% of all the KIF5B-RET mutations in our study. The prevalence of K15-R12 was higher than the predicted frequency of 75% in previous reports (9,13). CCDC6 was the second common fusion partner of RET, 8 patients were positive with CCDC6-RET fusion in our study. Besides fusing to RET, CCDC6 was also the fusion partner of PDGFR, ROS1, and KITLG in NSCLC samples. CCDC6 was necessary for the activation of RET and played a critical role in sustaining the DNA damage checkpoints. The defective of CCDC6 conferred the resistance to cis-platinum and sensitized cancer cells to small molecular inhibitors of repair enzymes (26,27). Several new or rare fusion partners of RET including ZNF438, ZNF43, STK33, REEP3, MRPS30, MIR7854, MARCH8, LOC401312, LINC01435, LINC00841, KIAA1217, ERC1, CBWD6, and ARHGAP12 was also identified in our study. NGS was a more comprehensive platform to detect the new fusion partners and concurrent gene mutation comparing to reverse transcription PCR (RT-PCR) and break-apart fluorescence in situ hybridization (FISH) (28). In our study, we applied the samples using the Lung Plasma panel and capture-targeted deep sequencing to better identify the RET fusion partners and concurrent gene mutations.
As an actionable driver mutation, RET fusions were considered to mutate exclusively with other actionable driver genes (29). However, we found concurrent TP53 and EGFR mutations in eight and three RET fusion positive patients, respectively. A 49-year-old female patient in our study was diagnosed with advanced (stage IV) LUAD, and received Bevacizumab combined with pemetrexed and AZD9291 (an EGFR inhibitor) as first-line and second-line treatment. After she was resistant to AZD9291, results of NGS suggested that the patient harbored ERC1-RET fusion and EGFR exon 19 mutation. Therefore, the patient was treated with Cabozantinib afterward. The efficacy was limited as the tumor progressed after 1 month. RET fusion might involve in the resistance of tumor cells with EGFR mutation to TKI therapy (13). The multi-target TKI Cabozantinib has been identified as a potent RET inhibitor. A phase II single-arm trial (NCT01639508) including 25 advanced NSCLC patients with RET rearrangement from the USA achieved 28% overall response and the median PFS and OS were 5.5 and 9.9 months respectively (30). Given that the overall response in this study was defined as the confirmed complete response or partial response, the overall response of Cabozantinib was 0. Although 39 patients was collected at the initiation of this study, Cabozantinib was administered in ten patients and only five samples in our study were available for the drug activity evaluation. Unfortunately, no significant survival benefit of patients receiving Cabozantinib was noted. Currently, most of the RET inhibitors were multi-targeted and some researches showed that the type of RET fusion might influence drug response (12). More precise and potent RET inhibitors may yield better clinical outcomes.
From the objective view, limitations were inevitably existed in our study. Firstly, the sample size of survival analysis was relatively small and the censoring rate was high which might contribute to the discrepancy between our results study and previous reports. Secondly, our study was designed as a retrospective analysis and some bias could not be avoided. In conclusion, RET fusion was a promising target in NSCLC, further randomized controlled clinical trials were warranted to validate the conclusion in the future.
Table S1
| RET fusion | Variant | N |
|---|---|---|
| KIF5B-RET | Exon 15–12 | 21 |
| Exon 15–13 | 1 | |
| Exon 16–12 | 1 | |
| Exon 23–12 | 1 | |
| CCDC6-RET | Exon 1–12 | 7 |
| Exon 2–12 | 1 | |
| ZNF438-RET | Intergenic region-exon 11 | 1 |
| ZNF43-RET | Intergenic region-exon 12 | 1 |
| STK33-RET | Exon 1–12 | 1 |
| REEP3-RET | Exon 5–12 | 1 |
| MRPS30-RET | Intergenic region-exon 12 | 1 |
| MIR7854-RET | Intergenic region-exon 12 | 1 |
| MARCH8-RET | Intergenic region-exon 12 | 1 |
| LOC401312-RET | Exon 2–12 | 1 |
| LINC01435-RET | Intergenic region-exon 12 | 1 |
| LINC00841-RET | Intergenic region-exon 12 | 1 |
| KIAA1217-RET | Intragenic region-exon12 | 1 |
| ERC1-RET | Exon 5–12 | 1 |
| CBWD6-RET | Exon 9–12 | 1 |
| ARHGAP12-RET | Exon 1–12 | 1 |
Acknowledgments
We thank Xinwei Zhang and Junling Li for financial support.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-754
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-754
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-754
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-754). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Research Ethics Committee of Cancer Hospital Chinese Academy of Medical Sciences (No.18-118/1696) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bethune G, Bethune D, Ridgway N, et al. Epidermal growth factor receptor (EGFR) in lung cancer: an overview and update. J Thorac Dis 2010;2:48-51. [PubMed]
- da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol 2011;6:49-69. [Crossref] [PubMed]
- Nagasaka M, Gadgeel SM. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev Anticancer Ther 2018;18:63-70. [Crossref] [PubMed]
- Mulligan LM. RET revisited: expanding the oncogenic portfolio. Nat Rev Cancer 2014;14:173-86. [Crossref] [PubMed]
- Gautschi O, Milia J, Filleron T, et al. Targeting RET in Patients With RET-Rearranged Lung Cancers: Results From the Global, Multicenter RET Registry. J Clin Oncol 2017;35:1403-10. [Crossref] [PubMed]
- Nelson-Taylor SK, Le AT, Yoo M, et al. Resistance to RET-Inhibition in RET-Rearranged NSCLC Is Mediated By Reactivation of RAS/MAPK Signaling. Mol Cancer Ther 2017;16:1623-33. [Crossref] [PubMed]
- Domvri K, Zarogoulidis P, Darwiche K, et al. Molecular Targeted Drugs and Biomarkers in NSCLC, the Evolving Role of Individualized Therapy. J Cancer 2013;4:736-54. [Crossref] [PubMed]
- Sculier JP, Berghmans T, Meert AP. Advances in target therapy in lung cancer. Eur Respir Rev 2015;24:23-9. [Crossref] [PubMed]
- Ferrara R, Auger N, Auclin E, et al. Clinical and Translational Implications of RET Rearrangements in Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:27-45. [Crossref] [PubMed]
- Drilon A, Wang L, Hasanovic A, et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013;3:630-5. [Crossref] [PubMed]
- Lee SH, Lee JK, Ahn MJ, et al. Vandetanib in pretreated patients with advanced non-small cell lung cancer-harboring RET rearrangement: a phase II clinical trial. Ann Oncol 2017;28:292-7. [Crossref] [PubMed]
- Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): an open-label, multicentre phase 2 trial. Lancet Respir Med 2017;5:42-50. [Crossref] [PubMed]
- Zhang K, Chen H, Wang Y, et al. Clinical Characteristics and Molecular Patterns of RET-Rearranged Lung Cancer in Chinese Patients. Oncol Res 2019;27:575-82. [Crossref] [PubMed]
- Inamura K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front Oncol 2017;7:193. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297-303. [Crossref] [PubMed]
- Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [Crossref] [PubMed]
- Newman AM, Bratman SV, Stehr H, et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390-3. [Crossref] [PubMed]
- Aboukhalil A, Bulyk ML. LOESS correction for length variation in gene set-based genomic sequence analysis. Bioinformatics 2012;28:1446-54. [Crossref] [PubMed]
- Yoo SS, Jin G, Jung HJ, et al. RET fusion genes in Korean non-small cell lung cancer. J Korean Med Sci 2013;28:1555-8. [Crossref] [PubMed]
- Lin C, Wang S, Xie W, et al. The RET fusion gene and its correlation with demographic and clinicopathological features of non-small cell lung cancer: a meta-analysis. Cancer Biol Ther 2015;16:1019-28. [Crossref] [PubMed]
- Tsuta K, Kohno T, Yoshida A, et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer 2014;110:1571-8. [Crossref] [PubMed]
- Michels S, Scheel AH, Scheffler M, et al. Clinicopathological Characteristics of RET Rearranged Lung Cancer in European Patients. J Thorac Oncol 2016;11:122-7. [Crossref] [PubMed]
- Gow CH, Liu YN, Li HY, et al. Oncogenic Function of a KIF5B-MET Fusion Variant in Non-Small Cell Lung Cancer. Neoplasia 2018;20:838-47. [Crossref] [PubMed]
- Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. [Crossref] [PubMed]
- Cerrato A, Merolla F, Morra F, et al. CCDC6: the identity of a protein known to be partner in fusion. Int J Cancer 2018;142:1300-8. [Crossref] [PubMed]
- Suzuki M, Makinoshima H, Matsumoto S, et al. Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci 2013;104:896-903. [Crossref] [PubMed]
- Drilon A, Hu ZI, Lai GGY, et al. Targeting RET-driven cancers: lessons from evolving preclinical and clinical landscapes. Nat Rev Clin Oncol 2018;15:151-67. [Crossref] [PubMed]
- Wu C, Zhao C, Yang Y, et al. High Discrepancy of Driver Mutations in Patients with NSCLC and Synchronous Multiple Lung Ground-Glass Nodules. J Thorac Oncol 2015;10:778-83. [Crossref] [PubMed]
- Drilon A, Rekhtman N, Arcila M, et al. Cabozantinib in patients with advanced RET-rearranged non-small-cell lung cancer: an open-label, single-centre, phase 2, single-arm trial. Lancet Oncol 2016;17:1653-60. [Crossref] [PubMed]