
ZC3H12D is a prognostic biomarker associated with immune cell infiltration in lung adenocarcinoma
Introduction
Primary lung cancer is one of the malignancies associated with the highest morbidity and mortality worldwide, especially in China (1). Non-small cell lung cancer (NSCLC) is the main pathologic type of lung cancer, accounting for approximately 80% of all lung cancers. Lung adenocarcinoma (LUAD) is the most common form of NSCLC, accounting for more than 50% of cases (2). The prognosis of LUAD is poor, with a 5-year survival rate of 4–17% (3). Immunotherapy is a new method for the treatment of LUAD, and has achieved unprecedented results in patients with advanced LUAD. However, there is a wide range of challenges in clinical application, such as the lack of biomarkers that can effectively predict efficacy, prognosis, and treatment-related adverse reactions (4). In addition, a number of studies have shown that tumor-infiltrating lymphocytes, such as tumor-associated macrophages (TAM) and tumor-infiltrating neutrophils (TINs), affect the prognosis and efficacy of immunotherapy (5). Therefore, the identification of prognostic biomarkers and new immune-related therapeutic targets has enormous potential in the treatment for LUAD.
ZC3H12 is a newly discovered zinc finger protein family of the CCCH type, which consists of four ZC3H12A-D members (6). The ZC3H12 protein family, also known as MCPIP, is an endonuclease that degrades mRNA transcripts by recognizing the stem-loop structure at the end of mRNA 3'UTR (7). ZC3H12 family proteins are abnormally expressed in a variety of tumors, and participate in the development and progression of tumors, thus affecting the prognosis. It has been reported that the expression of ZC3H12A is down-regulated in breast cancer, neuroblastomas, and renal clear cell carcinomas, and low expression of ZC3H12A is related to tumor grade and poor prognosis for breast cancer (8,9). Studies have shown that the expression of ZC3H12C in colon cancer is lower than normal colon tissues, while overexpression of ZC3H12C inhibits the migration of colon cancer cells (10). ZC3H12D was originally considered to be a tumor suppressor, and has also been referred to as p34, MCPIP4, and TFL (11). Recent studies have shown that ZC3H12D reduces the global ubiquitin of cellular proteins, and polyubiquitin plays a key role in promoting the tumor cell proliferation and survival signal pathway (12,13). It has also been reported that ZC3H12D is highly expressed in human endometrial carcinoma, but not expressed or weakly expressed in normal tissues, the patients with high expression of endometrial carcinoma have a good prognosis (14). Interestingly, members of the ZC3H12 family also provide a local inflammatory environment for tumor development. For example, ZC3H12A degrades transcripts of angiogenic factors secreted by cells (VEGF, IL-8, and IL-6) and downregulation of ZC3H12A induces tumor angiogenesis in renal cell carcinoma endothelial cells (8). Recently, ZC3H12D has been considered to be a new negative feedback regulator of Toll-like receptor (TLR) signaling and macrophage activation. Overexpression of ZC3H12D significantly inhibits JNK, ERK, and NF-κB signal transduction and macrophage inflammation induced by TLR2 and TLR4 activation (13). However, the mechanism of ZC3H12 family members in LUAD progression and tumor immunology has not been established.
In this study we used multiple databases to analyze the level of expression and prognosis of different ZC3H12 family proteins in patients with LUAD. We also used the tumor immune research evaluation resource (TIMER) to analyze the correlation between ZC3H12D and LUAD immune cell infiltration to determine the prognostic value of ZC3H12D in LUAD and its interaction with tumor immune potential.
Methods
UALCAN data analysis
UALCAN (http://ualcan.path.uab.edu) is an easy-to-use interactive portal for in-depth analysis of TCGA gene expression data. UALCAN can be used to analyze the relative expression of genes in different tumor types and grades, and to evaluate the impact of gene expression and clinicopathologic features on patient survival. In addition, additional information about the selected gene/target is provided through connection with HPRD, GeneCards, Pubmed, TargetScan, Human protein atlas, DRUGBANK, Open targets and GTEx. In this study, UALCAN database was used for the analysis of the expression of ZC3H12 family genes in primary LUAD patients and the investigation of the correlation between the ZC3H12 family members and the survival of patients.
Kaplan-Meier plotter database analysis
Kaplan-Meier Plotter analysis was used to assess how 10,461 cancer samples impacted 54,675 genes on survival. These samples included 5,143 breast, 1,816 ovarian, 2,437 lung, and 1,065 gastric cancer samples on the HGU133 Plus 2.0 array. The average follow-up time was 69, 40, 49 and 33 months, respectively (15). Kaplan-Meier Plotter (http://kmplot.com/analysis/) was used to analyze the relationship between the expression of ZC3H12 family genes and the survival of LUAD. The hazard ratio (HR) with 95% confidence intervals and log-rank P-value were also computed.
GEPIA was used to verify the correlation between ZC3H12 family genes and LUAD survival
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancerpku.cn/) is a web-based tool based on TCGA and GTEx data, including analysis of RNA sequencing expression data from 9,736 tumors and 8,587 normal samples from TCGA and GTEx (16). GEPIA provides key interactive analysis modules, including tumor/normal differential expression profile analysis, profile drawing, analysis based on tumor type or pathological stage, patient survival analysis, similar gene detection, correlation analysis and dimensionality reduction analysis, as well as rapid customization. We used GEPIA to further verify the correlation between ZC3H12 expression and clinical stage and survival of patients with LUAD.
Single cell sequencing analysis
CancerSEA (http://biocc.hrbmu.edu.cn/CancerSEA/http://202.97.205.69/CancerSEA/) is the first dedicated database designed to comprehensively explore the different functional states of cancer cells at the single cell level. CancerSEA mapped a single cell functional status map of cancer, including 14 functional states of 41,900 cancer cells from 25 cancer types (including stem cell, invasion, metastasis, proliferation, EMT, angiogenesis, apoptosis, cell cycle, differentiation, DNA damage, DNA repair, hypoxia, inflammation, and aging). CancerSEA facilitates determining which functional states are associated with genes of interest or a list of genes in different tumors (17). We used CancerSEA to analyze the correlation between ZC3H12 family genes and 14 functional states in LUAD.
TIMER database analysis
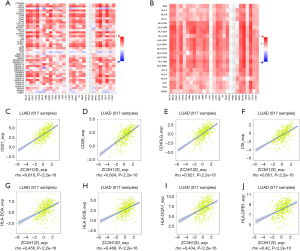
TIMER is a comprehensive database for systematic analysis of immune infiltration of different types of cancer (https://cistrome.shinyapps.io/timer/). TIMER used the previously published deconvolution statistical method to infer the abundance of tumor-infiltrating immune cells from gene expression profiles. The TIMER database included 10,897 samples from 32 cancer types from The Cancer Genome Atlas (TCGA) to estimate the abundance of immune infiltration. We analyzed the correlation between the expression of ZC3H12 family genes and the abundance of immune infiltrating cells (including B cells, CD4+T cells, CD8+T cells, neutrophils, macrophages, and dendritic cells) through a gene module. Gene expression against tumor purity is displayed on the left-most panel (18). In addition, the correlation between ZC3H12D expression and gene markers of tumor infiltrating immune cells was explored through related modules. The genetic markers of tumor infiltrating immune cells include CD8+T cells, T cell (general), B cells, monocytes, tumor associated macrophages, M1 macrophages, M2 macrophages, neutrophils, natural killer (NK) cells, dendritic cells (DC), T helper cell 1 (Th1) cells, T helper cell 2 (Th2) cells, follicular helper T cell (Tfh) cells. Helper T cell 17 (Th17), Tregs and T cell depletion). These genetic markers refer to previous studies. The correlation module generates expression scatter plots in a pair of user-defined genes, as well as Spearman's correlation and statistical significance. Log2 RSEM was used to show the level of gene expression.
TISIDB Database analysis
TISIDB (http://cis.hku.hk/TISIDB) is a user-friendly Web portal that gathers various types of tumor immunology data resources. It integrates 4,176 records from 2,530 publications, including 988 genes related to anti-tumor immunity. In addition, genes related to the resistance or sensitivity of tumor cells to T cell-mediated killing and immunotherapy were identified by analyzing high-throughput screening and genomic map data. Third, the association between any gene and immune function, such as lymphocytes, immunomodulators and chemokines, was calculated for 30 TCGA cancer types (19). We used TISIDB to analyze the role of ZC3H12D in the tumor-immune interaction.
Verification of ZC3H12D expression in GEO database
The gene expression profile was obtained from Gene Expression Omnibus (GEO) database. In this study, five GEO series (GSE) were used (GSE44077, GSE43458, GSE19804, GSE18842, and GSE19188). The dataset was selected as follows: the number of (I) samples >100 included normal and lung cancer tissues; and the number of samples in a single group of (II) was >30. The expression of ZC3H12D was analyzed using an unpaired t-test.
Immunohistochemical staining
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and it was conducted with approval from the Ethics Committee of Zhejiang Cancer Hospital (No. IRB-2015-176). Written informed consent was obtained from the participants. The Primary LUAD tissue and corresponding normal lung tissue used in this study were provided by the Biobank of Zhejiang Cancer Hospital. The primary antibody was a rabbit anti-human ZC3H12D polyclonal antibody purchased from Proteintech [24991-1-AP]. The immunohistochemical staining kit was purchased from Beijing Zhongshan Golden Bridge Biotechnology Co. Ltd. Paraffin-embedded tissue samples were sectioned and placed at 72 °C for 30 min, then dewaxed and hydrated by xylene and ethanol. Antigens were heat retrieved with citrate buffer. After three washes, the slides were placed in 3% hydrogen peroxide solution for 5–10 min and then washed with running water twice and PBS for 5 min. The primary antibodies were added (1:100 dilution) and incubated overnight at 4 °C. After washing with water twice and PBS for 5 min, the secondary antibodies were added and incubated for 20 min, followed by washing, DAB staining, counterstaining, and mounting.
Evaluation of immunohistochemical staining
The expression of ZC3H12D detected by immunohistochemistry was evaluated by measuring staining intensity and the proportion of positively-stained cells. The expression of ZC3H12D in LUAD is localized in the cytoplasm. According to the staining intensity, there were four levels including negative (0 point), weak positive (1 point), intermediate positive (2 points), and strong positive (3 points). The proportion of positive cells was scored as follows: <5% = 0 point, 5–25% = 1 point, 26–50% = 2 points, 51–75% = 3 points, and >75% = 4 points. If the sum of two scores was <4 points, it was considered low expression; 0 point indicated not expressed; and ≥4 points indicated high expression.
Statistical analysis
The statistical analysis survival curve was generated by UALCAN, Kaplan–Meier Plotter, and GEPIA. The results of UALCAN, Kaplan–Meier Plotter, and GEPIA are displayed with HR and P or Cox P-values from a log-rank test along with the values of HR and P or COX P from a logarithmic rank test. The correlation of gene expression was evaluated by Spearman correlation and statistical significance, and the following absolute thresholds were used to determine the intensity of the correlation: 0.00–0.19, “very weak”; 0.20–0.39, “weak”; 0.40–0.59, “medium”; 0.60–0.79, “strong”; and 0.80–1.0, “very strong.” P<0.05 was considered to be of statistical significance.
Results
Expression of ZC3H12 family members in LUAD
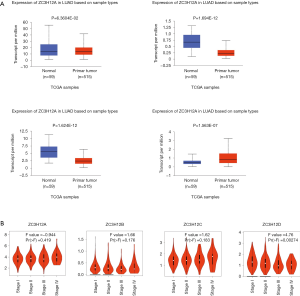
To determine the difference in ZC3H12 family member expression between LUAD and normal tissues, we used UALCAN to analyze the mRNA expression in ZC3H12 family members between LUAD and normal tissues. The results showed that ZC3H12A was slightly low in LUAD, but there was no statistical significance (P=0.0636). The expression of ZC3H12B and ZC3H12C was significantly decreased in LUAD, while the expression of ZC3H12D was significantly higher in LUAD (P<0.001; Figure 1A). At the same time, we used Expression DIY in GEPIA to analyze the expression of ZC3H12 family members in different pathologic stages of LUAD. The results showed that the expression of ZC3H12D decreased with progression of the disease [Pr (> F) = 0.00274], but there was no significant difference among the other three ZC3H12 family members (Figure 1B).
Prognostic significance of ZC3H12 family members in LUAD
We determined whether the expression of ZC3H12 family members was related to the prognosis of LUAD patients. First, the effect of ZC3H12 family member expression on the survival rate was evaluated by UALCAN. The prognosis of LUAD patients with high expression of ZC3H12D was significantly better than patients with low expression of LUAD (P=0.0048), while the expression of the other three genes was not significantly related to the prognosis of LUAD (Figure 2A,B,C,D). We used Kaplan–Meier Plotter to further evaluate the effect of ZC3H12 family member expression on the probability of survival among patients with LUAD. The data analysis based on RNA microarray showed that high expression of ZC3H12A was significantly correlated with poor prognosis (HR =1.75, 95% CI =1.38–2.21, P=3.3e-06). In contrast, the OS of patients with LUAD with high expression of the other three genes was significantly longer (ZC3H12B: HR =0.67, 95% CI =0.52–0.86, P=0.0018; ZC3H12C: HR =0.55, 95% CI =0.43–0.71, P=2.5e-06; and ZC3H12D: HR =0.52, 95% CI =0.41–0.67, P=3.6e-07; Figure 2E,F,G,H).
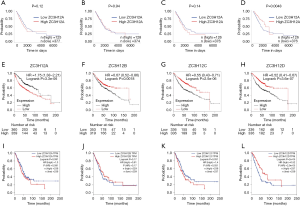
We further verified the prognostic significance of ZC3H12 family genes in patients with LUAD by using GEPIA. There was no correlation between the expression of ZC3H12A (HR =1.3, P=0.091) and ZC3H12B (HR =0.81, P=0.17) and OS in LUAD patients. High expression of ZC3H12C (HR =1.4, P=0.031) was significantly correlated with the poor prognosis of LUAD patients. In agreement with the survival data of UALCAN and the Kaplan–Meier Plotter, the high expression of ZC3H12D (HR =0.52, P=2.0e-05) was significantly correlated with the improvement in prognosis of patients with LUAD (Figure 2I,J,K,L). These results confirmed and supported the prognostic value of ZC3H12D in LUAD.
Functional state of ZC3H12 family members in LUAD
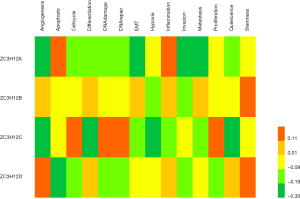
To better understand the role and potential mechanism underlying ZC3H12 expression in LUAD, we checked the functional status of ZC3H12 in LUAD in the CancerSEA database. The data on LUAD in CancerSEA came from the transcriptome (GSE69405) of individual cancer cells from LUAD patients with xenograft tumors (PDX). The data includes 3 datasets with a total of 126 single cells. Tumor cell-enriched PDX cells (LC-PT-45: scRNASeq, nasty 34) were analyzed and an additional batch (LC-Pt-45-Re: scRNA-Seq, nasty 43) was obtained to check comparable results. Another Lung cancer PDX case (LC-MBT15: scRNA-Seq, nasty 49) was prepared to validate our analytic strategy applied in the LC-PT-45 case. The results showed that there was no significant correlation between the ZC3H12 gene family and 14 functional states of LUAD (r<0.4; Figure 3).
Expression of ZC3H12D in LUAD is related to the level of immune infiltration
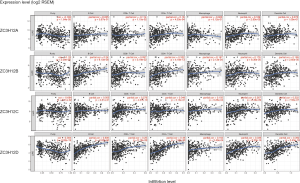
Tumor-infiltrating lymphocytes affect tumor development and patient survival. Therefore, we determined whether the expression of ZC3H12 family members was related to the level of immune infiltration in patient with LUAD. We evaluated the correlation between the expression of ZC3H12 family members and the level of immune infiltration. Among the four genes in the ZC3H12 family, with the exception of ZC3H12C, the expression of the other three genes was significantly correlated with tumor purity. Only the expression of ZC3H12D was positively correlated with the infiltration of B and CD4+ T cells (r>0.4, P<0.05; Figure 4). At the same time, we further verified the correlation between the expression of ZC3H12D in LUAD and immune cell infiltration by TISIDB. The expression of ZC3H12D was significantly correlated with activated and immature B cells (Figure S1). These results strongly suggested that ZC3H12D plays a special role in immune cell infiltration of LUAD, especially B cells.
Analysis of the correlation between ZC3H12D and the immune marker spectrum
To study the correlation between ZC3H12D and immune cell infiltration, we used TIMMER and GEPIA to analyze the correlation between ZC3H12D and a variety of immune cell markers in LUAD with ZC3H12A as a control. We analyzed the immune marker genes of ZC3H12D and different immune cells, including CD8+ T cells, T cells (general), B cells, monocytes, TAMs, M1 and M2 macrophages, neutrophils, NK cells and DCs with ZC3H12A as a control. At the same time, we also analyzed different functional T cells, such as Th1 cells, Th2 cells, Tfh cells, Th17 cells, Tregs, and depleted T cells (Table 1).
Table 1
| Description | Gene markers | ZC3H12D | ZC3H12A | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| None | Purity | None | Purity | |||||||
| Cor | P | Cor | P | Cor | P | Cor | P | |||
| CD8+ T cell | CD8A | 0.476 | *** | 0.376 | *** | 0.048 | 0.281 | −0.042 | 0.348 | |
| CD8B | 0.427 | *** | 0.348 | *** | 0.038 | 0.391 | −0.024 | 0.591 | ||
| CD3D | 0.572 | *** | 0.473 | *** | 0.122 | ** | 0.025 | 0.573 | ||
| CD3E | 0.631 | *** | 0.546 | *** | 0.106 | * | −0.005 | 0.907 | ||
| CD2 | 0.622 | *** | 0.533 | *** | 0.114 | ** | 0.011 | 0.806 | ||
| B cell | CD19 | 0.618 | *** | 0.546 | *** | 0.099 | * | 0.003 | 0.955 | |
| CD79A | 0.545 | *** | 0.462 | *** | 0.072 | 0.101 | −0.019 | 0.680 | ||
| Monocyte | CD86 | 0.453 | *** | 0.332 | *** | 0.088 | 0.0457 | 0.005 | 0.908 | |
| CSF1R | 0.46 | *** | 0.352 | *** | 0.131 | ** | 0.067 | 0.138 | ||
| TAM | CCL2 | 0.156 | *** | 0.019 | 0.669 | 0.199 | *** | 0.132 | ** | |
| CD68 | 0.3 | *** | 0.186 | *** | 0.111 | * | 0.042 | 0.351 | ||
| IL10 | 0.381 | *** | 0.265 | *** | 0.106 | * | 0.033 | 0.463 | ||
| Neutrophils | CEACAM8 | 0.135 | ** | 0.123 | ** | −0.06 | 0.171 | −0.064 | 0.154 | |
| ITGAM | 0.42 | *** | 0.32 | *** | 0.154 | *** | 0.093 | 0.0394 | ||
| CCR7 | 0.655 | *** | 0.576 | *** | 0.144 | ** | 0.05 | 0.268 | ||
| KIR2DL1 | 0.268 | *** | 0.224 | *** | 0.046 | 0.3 | 0.013 | 0.780 | ||
| KIR2DL3 | 0.234 | *** | 0.159 | *** | 0.074 | 0.0949 | 0.036 | 0.429 | ||
| KIR2DL4 | 0.24 | *** | 0.179 | *** | 0.115 | 0.0917 | 0.064 | 0.0833 | ||
| KIR3DL1 | 0.238 | *** | 0.181 | *** | 0.101 | 0.0213 | 0.075 | 0.0947 | ||
| KIR3DL2 | 0.324 | *** | 0.255 | *** | 0.026 | 0.553 | 0.017 | 0.661 | ||
| KIR3DL3 | 0.165 | *** | 0.15 | *** | 0.029 | 0.506 | 0.019 | 0.675 | ||
| KIR2DS4 | 0.243 | *** | 0.184 | *** | 0.031 | 0.476 | −0.001 | 0.985 | ||
| Dendritic cell | HLA-DPB1 | 0.457 | *** | 0.356 | *** | 0.098 | 0.0269 | 0.03 | 0.512 | |
| HLA-DQB1 | 0.3461 | *** | 0.241 | *** | 0.139 | ** | 0.084 | 0.0619 | ||
| HLA-DRA | 0.405 | *** | 0.293 | *** | 0.082 | 0.0639 | 0.013 | 0.773 | ||
| HLA-DPA1 | 0.418 | *** | 0.318 | *** | 0.057 | 0.195 | −0.009 | 0.841 | ||
| CD1C | 0.329 | *** | 0.239 | *** | 0.076 | 0.0848 | 0.024 | 0.593 | ||
| NRP1 | 0.096 | * | 0.047 | 0.293 | −0.024 | 0.586 | −0.047 | 0.300 | ||
| CD11c | 0.54 | *** | 0.455 | *** | 0.28 | *** | 0.224 | *** | ||
| Th1 | TBX21 | 0.544 | *** | 0.457 | *** | 0.139 | ** | 0.046 | 0.306 | |
| STAT4 | 0.428 | *** | 0.316 | *** | 0.204 | *** | 0.119 | ** | ||
| STAT1 | 0.244 | 0.121 | 0.14 | ** | 0.057 | 0.195 | −0.012 | 0.784 | ||
| IFNG | 0.35 | * | 0.263 | *** | 0.123 | ** | 0.066 | 0.143 | ||
| TNF-α | 0.446 | *** | 0.335 | *** | 0.086 | * | 0.297 | *** | ||
| Th2 | GATA3 | 0.333 | *** | 0.202 | *** | 0.086 | * | −0.006 | 0.889 | |
| STAT6 | 0.195 | *** | 0.217 | *** | 0.179 | *** | 0.176 | *** | ||
| STAT5A | 0.5 | *** | 0.396 | *** | 0.183 | *** | 0.117 | *** | ||
| IL13 | 0.191 | *** | 0.123 | ** | 0.183 | *** | 0.117 | ** | ||
| Tfh | BCL6 | 0.121 | ** | 0.129 | ** | 0.183 | *** | 0.14 | ** | |
| IL21 | 0.35 | *** | 0.303 | *** | 0.028 | 0.523 | −0.008 | 0.857 | ||
| Th17 | STAT3 | 0.141 | ** | 0.162 | *** | −0.056 | 0.206 | −0.05 | 0.270 | |
| IL17A | 0.235 | *** | 0.17 | *** | 0.17 | *** | 0.14 | ** | ||
| Treg | FOXP3 | 0.573 | *** | 0.481 | *** | 0.146 | *** | 0.047 | 0.300 | |
| CCR8 | 0.589 | *** | 0.501 | *** | 0.063 | 0.153 | −0.025 | 0.577 | ||
| STAT5B | 0.416 | *** | 0.409 | 0.593 | −0.044 | 0.316 | −0.06 | 0.183 | ||
| TGFB1 | 0.245 | *** | 0.142 | *** | 0.159 | *** | 0.099 | * | ||
| Mast | TLR2 | 0.254 | *** | 0.169 | *** | 0.08 | 0.0713 | 0.0464 | 0.304 | |
| TLR4 | 0.441 | *** | 0.322 | *** | 0.009 | 0.843 | −0.08 | 0.0776 | ||
| TPSB2 | 0.191 | *** | 0.078 | 0.0839 | 0.131 | ** | −0.225 | *** | ||
| CD33 | 0.423 | *** | 0.312 | *** | 0.119 | ** | 0.054 | 0.231 | ||
| CD44 | 0.214 | *** | 0.085 | *** | 0.123 | ** | 0.07 | 0.119 | ||
| T cell exhaustion | PDCD1 | 0.536 | *** | 0.454 | ** | 0.212 | *** | 0.144 | ** | |
| CTLA4 | 0.632 | *** | 0.558 | *** | 0.229 | *** | 0.154 | *** | ||
| LAG3 | 0.426 | *** | 0.348 | *** | 0.18 | *** | 0.128 | ** | ||
| HAVCR2 | 0.408 | *** | 0.281 | *** | 0.11 | * | 0.026 | 0.560 | ||
| GZMB | 0.344 | *** | 0.243 | *** | 0.157 | 0.815 | 0.097 | * | ||
*, P<0.05; **, P<0.01; ***, P<0.001.
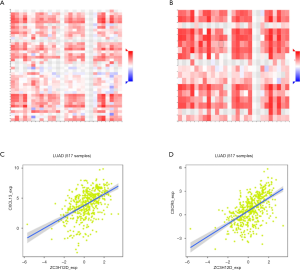
Interestingly, most of the B cell markers were associated with ZC3H12D expression. Specifically, the expression of ZC3H12D was significantly correlated with the B cell markers, CD79 and CD19 (P<0.001). We used GEPIA to verify the correlation between ZC3H12D and the aforementioned markers (Table 2). At the same time, we also analyzed the correlation between the expression of ZC3H12D and immunoinhibitors, immunostimulators, and MHC molecules. The expression of ZC3H12D was positively correlated with immunostimulators, including CD27, CD28, CD40LG, and LTA, and MHC molecules, including HLA-DOA, HLA-DOB, HLA-DQA1, and HLA-DPB1 (Figure 5). In addition, the correlation between ZC3H12D and immune-related chemokines and receptors was evaluated. The expression of ZC3H12D was significantly correlated with the chemokine, CXCL13, and its receptor (CXCR5) derived from B cells (Figure 6).
Table 2
| Description | Gene marker | ZC3H12D | ZC3H12A | |||
|---|---|---|---|---|---|---|
| R | P | R | P | |||
| CD8+ T cell | CD8A | 0.44 | *** | 0.032 | 0.48 | |
| CD8B | 0.38 | *** | −0.0039 | 0.93 | ||
| CD3D | 0.55 | *** | 0.086 | 0.059 | ||
| CD3E | 0.62 | *** | 0.1 | * | ||
| CD2 | 0.62 | *** | 0.094 | * | ||
| B cell | CD19 | 0.58 | *** | 0.11 | * | |
| CD79A | 0.48 | *** | 0.027 | 0.55 | ||
*, P<0.05; ***, P<0.001.
ZC3H12D expression verification
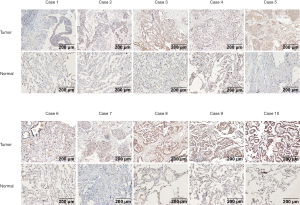
The datasets that met the criteria were filtered through the GEO database. There were five datasets that met the criteria, as follows: GSE18842; GSE19188; GSE19804; GSE43458; and GSE44077. There were 332 cases of NSCLC and 266 normal controls. The results of a non-paired t-test showed that the expression of ZC3H12D was significantly higher in 286 cases of NSCLC in the GSE19188, GSE19804, GSE43458, and GSE44077 datasets than 221 cases in the control group, while ZC3H12D was highly expressed in tumor groups in the GSE18842 dataset, but the difference was not statistically significant (Figure S2). We then analyzed 10 pairs of LUAD tissues and the corresponding normal lung tissues to verify ZC3H12D expression. IHC staining showed that ZC3H12D was abundantly and uniformly expressed in tumor samples but was significantly down-regulated in all adjacent non-tumor lung tissues (Figure 7). The clinicopathological information of patients with LUAD used in immunohistochemical is shown in Table S1. In general, like the TCGA database based on RNA high-throughput sequencing, ZC3H12D was also highly expressed in chip-based datasets and was highly expressed in LUAD tissues, which can be used as a marker of LUAD prognosis and immune cell infiltration.
Discussion
The relationship between ZC3H12 family dysfunction and a variety of cancers has been partially reported (20-22); however, the biological analysis of the ZC3H12 family in LUAD has not been reported. This study analyzed the expression and prognosis of ZC3H12 family members in LUAD. ZC3H12D was significantly up-regulated in early LUAD tissues, and the patients with high expression of LUAD had a good prognosis. Independent databases (UALCAN, Kaplan-Meier Plotter and GEPIA analysis) were used to analyze the prognosis of ZC3H12 family members in patients with LUAD. Due to the difference in number of LUAD patients included in the survival statistics and the threshold of high and low classification, the results of the databases were different, but together the three databases showed that there was a positive correlation and consistency between ZC3H12D and LUAD prognosis. In addition, we also found that there was no significant correlation between ZC3H12D and 14 functional states of the tumor. Our results provide important insight into the lack of ZC3H12D expression as a poor prognostic marker in LUAD.
Another important finding in this study was that the expression of ZC3H12D was significantly correlated with the level of LUAD B cell infiltration and CD4+T cell infiltration. The correlation between ZC3H12D expression and immunocyte marker genes suggests that ZC3H12D plays an important role in tumor immune regulation of LUAD. Specifically, ZC3H12D was significantly correlated with the B cell markers, CD79 and CD19. The expression of ZC3H12D was positively correlated with immunostimulators, including CD27, CD28, CD40LG, and LTA, and MHC molecules, including HLA-DOA, HLA-DOB, HLA-DQA1, and HLA-DPB1. The expression of ZC3H12D was positively correlated with cytokines, including TNF and LTA, CXCL13, and its receptor CXCR5.
Recent studies have explained the possible mechanism of ZC3H12D expression associated with immune infiltration and good prognosis. Tumor-infiltrating B cells (TIBs) are important regulatory cells in the progression of lung cancer and has a dual role in the tumor microenvironment (23). It has been reported that the number of TIBs is positively correlated with the prognosis of NSCLC, especially in LUAD (24). TIBs promote T cell responses by secreting cytokines and chemokines, maintains the neutralization function of the tertiary lymphoid structure (TLS) of lung cancer, and has an anti-tumor immune effect (25). For example, B cells produce CXCL13 and lymphotoxin (LT) through TLR4 signals. Moreover, TLR4 signals are positive feedback loops to support TLS formation and high density (26). Some studies have shown that CXC13-coupled CpG-ODN stimulation of CXCR5-expressing B cells trigger the killing effect of CD8+T cells, thus eliminating lung metastasis in 4T1.2 tumor-bearing mice (27). B cells with a tumor-promoting effect are defined as regulatory B cells (Bregs). Bregs secreting cytokines, such as IL-10 and TGF-β, inhibit Th1/CD8+T cell function and promote tumor proliferation (28). Some Bregs have a CD19+CD5+ phenotype driven by STAT3 activation and overexpression of CD5 induced by IL-6, which is thought to promote tumor progression by inducing tumor angiogenesis and immunity (29). Some studies have shown that ZC3H12D is involved in the degradation of pro-inflammatory mRNA, such as mRNA of IL-6, IL-10, and IER3 (30). ZC3H12D leads to a decrease in IL-10 and IL-6, inhibits Bregs, and exhibits an anti-tumor effect.
In conclusion, the increase of ZC3H12D expression is related to the good prognosis of LUAD and the increase in B cells. Therefore, ZC3H12D may play an important role in LUAD immune cell infiltration as a biomarker of infiltration and prognosis.
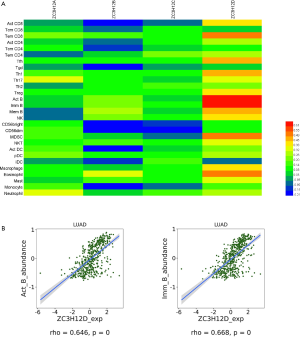
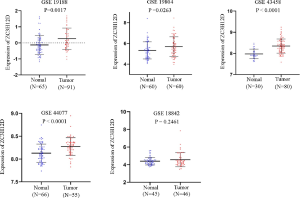
Table S1
| Cases | Gender | Age, years | Tumor diameter, cm | Lymph node metastasis | Smoking index | Differentiation | Vascular tumor thrombus | Visceral pleura involvement |
|---|---|---|---|---|---|---|---|---|
| 1 | Female | 62 | 6×5×4.5 | Yes | 0 | Moderately-low | No | No |
| 2 | Female | 50 | 4.5×3.7×2.6 | Yes | 0 | – | Yes | Yes |
| 3 | Female | 53 | 2.3×2×2 | No | 0 | – | No | Yes |
| 4 | Female | 64 | 2.7×2×1.5 | Yes | 0 | – | Yes | Yes |
| 5 | Male | 52 | 7×6×4 | Yes | >20 | Moderately-low | Yes | Yes |
| 6 | Female | 79 | 2×1.8×1.5 | Yes | >20 | – | No | Yes |
| 7 | Male | 58 | 3.3×2.5×2 | No | >20 | Moderately-low | Yes | No |
| 8 | Male | 66 | 4×3.8×2.4 | Yes | >20 | – | No | Yes |
| 9 | Male | 53 | 2×1.5×1 | Yes | >20 | Moderately-low | Yes | Yes |
| 10 | Female | 59 | 2×1.5×1 | No | 0 | Moderately | No | Yes |
Acknowledgments
Funding: Our study was supported financially by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1465
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1465). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki and it was conducted with approval from the Ethics Committee of Zhejiang Cancer Hospital (No. IRB-2015-176). Written informed consent was obtained from the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Harada H, Murayama S. Proton beam therapy in non-small cell lung cancer: state of the art. Lung Cancer (Auckl) 2017;8:141-5. [Crossref] [PubMed]
- Camidge DR, Doebele RC, Kerr KM. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nat Rev Clin Oncol 2019;16:341-55. [Crossref] [PubMed]
- Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 2020;20:25-39. [Crossref] [PubMed]
- Liang J, Wang J, Azfer A, et al. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J Biol Chem 2008;283:6337-46. [Crossref] [PubMed]
- Fu M, Blackshear PJ. RNA-binding proteins in immune regulation: a focus on CCCH zinc finger proteins. Nat Rev Immunol 2017;17:130-43. [Crossref] [PubMed]
- Marona P, Gorka J, Mazurek Z, et al. MCPIP1 Downregulation in Clear Cell Renal Cell Carcinoma Promotes Vascularization and Metastatic Progression. Cancer Res 2017;77:4905-20. [Crossref] [PubMed]
- Lu W, Ning H, Gu L, et al. MCPIP1 Selectively Destabilizes Transcripts Associated with an Antiapoptotic Gene Expression Program in Breast Cancer Cells That Can Elicit Complete Tumor Regression. Cancer Res 2016;76:1429-40. [Crossref] [PubMed]
- Suk FM, Chang CC, Lin RJ, et al. MCPIP3 as a Potential Metastasis Suppressor Gene in Human Colorectal Cancer. Int J Mol Sci 2018;19:1350. [Crossref] [PubMed]
- Wang M, Vikis HG, Wang Y, et al. Identification of a novel tumor suppressor gene p34 on human chromosome 6q25.1. Cancer Res 2007;67:93-9. [Crossref] [PubMed]
- Chan CH, Jo U, Kohrman A, et al. Posttranslational regulation of Akt in human cancer. Cell Biosci 2014;4:59. [Crossref] [PubMed]
- Huang S, Qi D, Liang J, et al. The putative tumor suppressor Zc3h12d modulates toll-like receptor signaling in macrophages. Cell Signal 2012;24:569-76. [Crossref] [PubMed]
- Wakahashi S, Kawakami F, Wakahashi K, et al. Transformed Follicular Lymphoma (TFL) Predicts Outcome in Advanced Endometrial Cancer. Cancer Epidemiol Biomarkers Prev 2018;27:963-9. [Crossref] [PubMed]
- Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer 2012;19:197-208. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Yuan H, Yan M, Zhang G, et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res 2019;47:D900-8. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 2019;35:4200-2. [Crossref] [PubMed]
- Ligeza J, Marona P, Gach N, et al. MCPIP1 contributes to clear cell renal cell carcinomas development. Angiogenesis 2017;20:325-40. [Crossref] [PubMed]
- Labedz-Maslowska A, Lipert B, Berdecka D, et al. Monocyte Chemoattractant Protein-Induced Protein 1 (MCPIP1) Enhances Angiogenic and Cardiomyogenic Potential of Murine Bone Marrow-Derived Mesenchymal Stem Cells. PloS One 2015;10:e0133746. [Crossref] [PubMed]
- Skalniak A, Boratyn E, Tyrkalska SD, et al. Expression of the monocyte chemotactic protein-1-induced protein 1 decreases human neuroblastoma cell survival. Oncol Rep 2014;31:2385-92. [Crossref] [PubMed]
- Patel AJ, Richter A, Drayson MT, et al. The role of B lymphocytes in the immuno-biology of non-small-cell lung cancer. Cancer Immunol Immunother 2020;69:325-42. [Crossref] [PubMed]
- Kinoshita T, Muramatsu R, Fujita T, et al. Prognostic value of tumor-infiltrating lymphocytes differs depending on histological type and smoking habit in completely resected non-small-cell lung cancer. Ann Oncol 2016;27:2117-23. [Crossref] [PubMed]
- Wang SS, Liu W, Ly D, et al. Tumor-infiltrating B cells: their role and application in anti-tumor immunity in lung cancer. Cell Mol Immunol 2019;16:6-18. [Crossref] [PubMed]
- Litsiou E, Semitekolou M, Galani IE, et al. CXCL13 production in B cells via Toll-like receptor/lymphotoxin receptor signaling is involved in lymphoid neogenesis in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2013;187:1194-202. [Crossref] [PubMed]
- Bodogai M, Lee Chang C, Wejksza K, et al. Anti-CD20 antibody promotes cancer escape via enrichment of tumor-evoked regulatory B cells expressing low levels of CD20 and CD137L. Cancer Res 2013;73:2127-38. [Crossref] [PubMed]
- Taghavi N, Mohsenifar Z, Baghban AA, et al. CD20+ Tumor Infiltrating B Lymphocyte in Oral Squamous Cell Carcinoma: Correlation with Clinicopathologic Characteristics and Heat Shock Protein 70 Expression. Patholog Res Int 2018;2018:4810751. [Crossref] [PubMed]
- Zhang C, Xin H, Zhang W, et al. CD5 Binds to Interleukin-6 and Induces a Feed-Forward Loop with the Transcription Factor STAT3 in B Cells to Promote Cancer. Immunity 2016;44:913-23. [Crossref] [PubMed]
- Wawro M, Kochan J, Krzanik S, et al. Intact NYN/PIN-Like Domain is Crucial for the Degradation of Inflammation-Related Transcripts by ZC3H12D. J Cell Biochem 2017;118:487-98. [Crossref] [PubMed]


