Genomic approach to translational studies in colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most common malignant neoplasm worldwide and a leading cause of cancer-related morbidity and mortality (1). Metastatic CRC (also called stage IV or advanced CRC) is the principal cause of death, but if cancer is detected at early stages curative treatment is often possible. Surgery is the primary form of treatment and results in cure for ~60% of patients with localized (stage I-III) disease (2,3). However, recurrence following surgery remains a major problem, and patients with lymph-node positive stage III and high-risk stage II disease are offered fluoropyrimidine-based adjuvant chemotherapy (5-fluorouracil, capecitabine) with or without oxaliplatin. Rectal cancers may also receive pre-operative chemoradiation. Recurrent tumor can develop in the bowel or at distant sites including the liver, lung, peritoneum, brain and bone (2,3). Guidelines for post-surgery surveillance recommend a combination of clinical assessment, serum carcinoembryonic antigen (CEA) testing, colonoscopy and computed tomography (CT) scanning (4-6). In current practice, many CRC patients receive adjuvant therapy unnecessarily, either because they were cured by surgery alone, or because they will relapse despite treatment. Conversely, some stage II patients with low-risk clinicopathological features who are currently not considered for adjuvant therapy do relapse and might benefit from therapy.
Outcomes from metastatic CRC remain poor, with a 5-year survival rate of less than 20% (7). Curative surgery is only rarely possible in these patients, but an increase in therapeutic options has resulted in an improvement of median overall survival to ~24 months. Approved agents include standard chemotherapeutics (5-fluorouracil, capecitabine, oxaliplatin, irinotecan) and targeted therapies directed against the epidermal growth factor receptor (EGFR) (cetuximab, panitumumab) or angiogenesis (bevacizumab, aflibercept, regorafenib). Although these treatments have prolonged the lives of patients with metastatic CRC, clinical responses are limited to a subset of individuals and are generally short-lived with most tumors developing resistance within a few months. Significant side effects and costs are associated with these treatments, and identification of individuals who are likely to derive the greatest benefit remains a major challenge.
Many patients with CRC will remain asymptomatic until the development of late-stage disease, where symptoms may include abdominal pain, changes in bowel habit and the presence of blood in stool. The principal method adopted by national CRC screening programs for early disease detection is the fecal occult blood test (FOBT), targeted at high-risk age groups with follow-up by colonoscopy (8,9). FOBT screening is cost effective, but tests suffer from limited sensitivity and specificity. A further challenge is population participation for stool-based diagnostics.
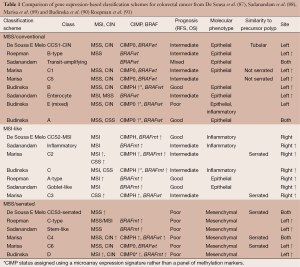
Advances in the development of microarray and next-generation sequencing (NGS) technologies have enabled global studies of CRC genomes, methylomes, as well as coding and non-coding transcriptomes (Figure 1). Integrated omics data have led to the identification of new cancer genes and pathways, and have improved our understanding of tumor biology and molecular subtypes. Translational genomics studies have revealed clinically relevant biomarkers for improving CRC diagnosis, surveillance, prediction of prognosis and therapy response. In addition, such studies have identified new druggable targets, opening up novel therapeutic opportunities. Here, we summarize pertinent results of CRC genomics studies to date, with an emphasis on tumor classification, diagnosis, prognostication and prediction of therapy benefit.
The CRC genome
Comparative genomic hybridization (CGH) arrays, single nucleotide polymorphism (SNP) arrays and more recently NGS approaches have provided fundamental insights into the complex landscapes of CRC mutations, DNA copy number alterations and chromosomal rearrangements. Sjöblom et al. and Wood et al. first used classic PCR-based Sanger sequencing for exome-wide profiling of CRC mutations, identifying well-known, high-frequency mutated genes such as APC, KRAS, PIK3CA, SMAD4, TP53 and FBXW7 as ‘gene mountains’, and describing a large number of ‘gene hills’ that were mutated at low frequency (10,11). These pioneering studies were followed by integrated whole-exome NGS and DNA copy-number studies by The Cancer Genome Atlas (TCGA) Network, presenting a detailed survey of the genomic profiles on over 270 sporadic CRCs (12). Approximately 15% of CRCs were found to exhibit hypermutation with two distinct mutation patterns: microsatellite instability (MSI) in three-quarters of cases, characterized by increased insertions, deletions and single nucleotide substitutions, usually with hypermethylation and MLH1 silencing, and a nucleotide substitution hypermutator phenotype (NSHP) in one-quarter of cases, associated with mutations in polymerase ε (POLE). Twenty-four genes were highlighted as significantly mutated, targeting the WNT, RTK/RAS, PI3K, TGF-β and TP53 pathways in both non-hypermutated and hypermutated tumors, but with different genetic alterations between these CRC subtypes. Non-hypermutated tumors showed common mutations in APC, TP53, KRAS, PIK3CA, FBXW7, SMAD4, TCF7L2, NRAS, CTNNB1, SMAD2, FAM123B, SOX9, ATM and ARID1A, while hypermutated tumors showed frequent alterations in ACVR2A, APC, TGFBR2, BRAF, MSH3, MSH6, SLC9A9 and TCF7L2. At the chromosomal level, non-hypermutated tumors tended to be aneuploid, while hypermutated tumors tended to be near-diploid. Consistent with previous CGH and SNP array studies (13-18), the most commonly deleted chromosome arms were 8p, 15q, 17p (including TP53) and 18q (including SMAD4), and the most commonly gained regions were chromosome 7, 8q (including MYC), 13 and 20q. Recurrent copy-number alterations included potentially drug-targetable amplifications of ERBB2 and IGF2. Low prevalence chromosomal translocations were detected between NAV2 and the WNT pathway member TCF7L1 using whole-genome sequencing on a subset of samples. A similar genomic study on 74 primary colon tumors reported highly concordant results, and also identified recurrent fusion transcripts involving R-spondin family members (EIF3E-RSPO2 and PTPRK-RSPO3) that were shown to contribute to activation of oncogenic WNT/β-catenin signaling (19,20). Additional low prevalence translocations identified by whole-genome or targeted NGS studies in CRC include C2orf44-ALK, VTI1A-TCF7L2 and LACTB2-NCOA2 (19,21,22). Recent NGS studies have provided additional details on the mutation spectra of colorectal adenomas, MSI and microsatellite stable (MSS) carcinomas (23-29). Mutational heterogeneity has been investigated between primary cancers and matched metastases indicating high genomic concordance, with a thick common trunk and smaller genomic branches (30-33). Some evidence exists for intra-tumor mutational heterogeneity, but data on this are still emerging (33). CGH array studies have proposed a refined classification of non-hypermutated CRCs into chromosomally stable (CSS) and chromosomal instability (CIN) groups (17,34). However, these groups have not as yet been systematically investigated for specific mutation signatures.
Non-invasive analysis of circulating tumor DNA (ctDNA) is an emerging genomics-tool that is actively being developed to improve CRC diagnosis and post-surgery surveillance. It is based on the detection of tumor specific single-base substitutions or larger somatic structural variations (SSVs) in DNA fragments that are released by tumors into plasma. Assays are typically designed against either point mutations in hotspot genes or patient-specific SSVs (35-40). Hotspot mutations can be utilized in both the diagnostic and surveillance setting, but these may only identify a subset of patients and have limited specificity. Application of patient-specific SSVs is restricted to the surveillance setting, requiring low coverage whole-genome sequencing and/or microarray analysis of resected tumor for assay design, yet highly-specific tests can in principle be produced for all individuals. Several reports have shown that assays against point mutations in hotspot mutated genes like KRAS, BRAF and PIK3CA can identify ctDNA fragments in plasma and serum in ~70% of patients with CRC (38,39). Recently, a clinical pipeline for identification of patient-specific SSVs for post-surgery CRC surveillance has been presented, demonstrating sensitive temporal assessment of disease status, response to surgical and oncological intervention, and early detection of recurrence (40). Recommending the use of at least three SSVs per patient to counter observed primary-metastasis genetic heterogeneity, this approach achieved sensitivity and specificity of 100% for detecting relapse, with a 2-15 (mean 10) months lead time compared to conventional follow-up.
Stool-based diagnostic tests have also been successfully tested for detection of mutations in high-frequency mutated CRC genes, including APC, KRAS and TP53 (41-48) such as the clinically used PreGen-Plus™ kit (49). Additionally, studies have evaluated long fragment DNA from exfoliated cancer cells in stool as diagnostic marker, with modest sensitivity and specificity (50-52).
Genomic instability phenotypes of CRC, MSI and CIN, have been demonstrated to be predictive of good and poor prognosis, respectively (53,54). The extent of CIN may provide additional prognostic value (55,56). Several CGH array studies have attempted to define particular regions of chromosomal gain or loss related to tumor progression and outcome (14,16,57-68). Perhaps the strongest data exist for loss of chromosome arms 4q and 18q and inferior survival, but whether these relationships are independent of global CIN status remains uncertain (68,69). Recently, different types of CIN, such as genome-doubling and chromothripsis, have been suggested to be adversely related with patient outcome (70,71).
Targeted gene sequencing studies to develop integrated mutation signatures for CRC prognostication are only beginning to emerge. A recent study evaluating 187 recurrent and pathway-related genes in 160 patients with stage I-IV CRCs, has proposed a five-gene-signature (CDH10, COL6A3, SMAD4, TMEM132D, VCAN) for stratifying patients by outcome independent of TNM status (72).
Genomic approaches are gradually being applied for identification of molecular markers of therapy benefit. To date, unbiased exome mutation and DNA copy number studies have focused on cancer cell lines in the context of high-throughput drug screens. However, only small numbers of CRC cell lines have been included in such screens thus limiting the power of these studies to identify robust biomarker-drug response associations (73-75). In patients with metastatic CRC, several targeted gene mutation and copy-number analyses have investigated resistance to treatment with monoclonal antibodies targeting EGFR. These studies have largely considered “rational” candidate genes indicated by previous focused studies. For example, Peeters et al. evaluated cancer resistance to panitumumab using massively parallel multigene tumor sequencing of KRAS, NRAS, BRAF, PIK3CA, PTEN, TP53, EGFR, AKT1 and CTNNB1. As found in other reports (76,77), wild-type KRAS, NRAS and BRAF status were associated with longer progression-free survival (78). Ciardiello et al. reported a similar targeted NGS study interrogating 22 genes in patients treated with FOLFIRI plus cetuximab, reporting worse outcome in cases with KRAS, NRAS, BRAF, or PIK3CA mutations (79). The potential of ctDNA analysis for monitoring intrinsic and acquired resistance to anti-EGFR antibody therapy has been successfully demonstrated, applying both targeted mutation and SSV analysis (38,80-82). Limited data suggest that tumor DNA copy number profiles may correlate with outcome in advanced CRC patients treated with fluoropyrimidine-based regimens. In particular, chromosomal losses of 18q, 17p11.2-p13.2 and gains of 20p13-q13.3 have been associated with response to the FU + irinotecan (FOLFIRI) and capecitabine + irinotecan (CAPIRI) (83,84).
Limited data exist for rectal cancer response to preoperative chemoradiation. A study by Chen et al. highlighted loss of chromosome 4 as associated with the risk of lymph node metastasis (85). Similarly, Grade et al. suggested that pre-therapeutic evaluation of gains of chromosomal regions 7q32-q36 and 7q11-q31, and amplifications of 20q11-q13 may predict responsiveness to chemoradiotherapy (86).
The CRC transcriptome
Analysis of the protein-coding CRC transcriptome using microarray platforms has provided a framework for classification of CRC subtypes and prediction of cancer outcomes and therapy benefit. These signatures are generally derived from the analysis of resected tumor specimens with limited micro-dissection and capture neoplastic, stromal and immune components.
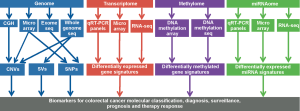
Several classification schemes for CRC have been proposed based on unsupervised clustering of tumor gene expression data (87-91). Approaches to tumor categorization have included hierarchical clustering, non-negative matrix factorization and the clustering of meta-genes (medians of groups of genes with correlated expression). Although these classification schemes differ in the number and detail of the subtypes proposed, ranging from three to six groups, major themes are separation into classes differentiated by MSI and CIN status, tumor location, and expression of epithelial versus mesenchymal markers (Table 1). One study has aligned their classification with different types of precursor lesions, classic versus serrated adenoma (87), while another has connected their classes with the cell types in colorectal crypts, stem cell, transit-amplifying cell, goblet cell and enterocyte (88). Recently, two studies have demonstrated major contributions of stromal cells in tumor groups with increased mesenchymal marker expression, rather than tumor cells undergoing epithelial-to-mesenchymal transition as was originally proposed (92,93).
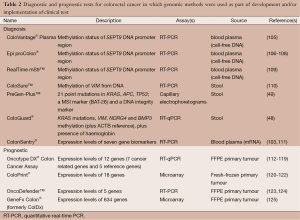
mRNA extracted from blood and stool have been considered as biomarker analysis for diagnosis of CRC (94-96). Several groups have used expression microarrays on blood from patients with CRC and healthy controls to identify an initial set of candidate diagnostic mRNAs followed by further refinement of candidates using RT-PCR (96,97). Other groups have screened normal and tumour tissue to find differentially expressed candidate genes, from which a refined set was obtained upon follow-up using mRNA extracted from stool-derived colonocytes (98) or blood (99). A related approach has been to examine previously described candidate genes with reported high levels of expression in tumour or patient blood (e.g., COX2, MMP7 and CEA) (100-102). One commercial blood-based diagnostic test for CRC, ColonSentry®, which uses a 7-gene mRNA signature is currently available (103,104) (Table 2). Validation in two cohorts yielded sensitivity and specificity of 72-82% and 64-70%, respectively (103,111). However, research into using mRNA for CRC diagnosis appears to be waning relative to approaches which utilise aberrant DNA methylation or miRNAs discussed below.
Full table
Multiple studies (126-139) have searched for gene expression signatures for predicting risk of tumor recurrence following surgical resection of the primary tumor (112,120,125,140-155). Early studies often had modest sample sizes and relied on cross-validation to assess performance of their signatures, while later studies evaluated larger sample sizes and included independent patient cohorts for signature assessment. A survey of 31 gene signatures demonstrated little overlap in the component genes (156), and only modest prognostic performance when assessed in independent datasets (156). Recognized reasons for these findings are technical differences in sample preparation and microarray processing, cohort heterogeneity and gene selection methods. These challenges can be overcome using rigorously controlled assay conditions. Accordingly, four prognostic gene expression signatures have been translated into clinical use following extensive validation on external cohorts: Oncotype DX® (Colon), a 12-gene RT-PCR based assay (112-119,157), ColoPrint®, an 18 gene microarray-based assay (120-122), OncoDefender™, a 5 gene RT-PCR based assay (123,124), and GeneFx Colon®, a 634 gene microarray-based assay (125) (Table 2). Gene expression based CRC classification schemes may also have prognostic potential, with MSI-associated classes showing good prognosis and serrated/mesenchymal classes exhibiting poor prognosis (87-91).
Transcriptome analyses have also been attempted for predicting response to chemotherapy and radiotherapy for CRC. Perhaps the most studied scenario has been that of pre-operative chemoradiation in rectal cancer patients, utilizing pre-treatment biopsies (158-165). These transcriptomic studies typically involve smaller training sets (n<100) than those for prognosis signatures and generally lack external validation. Classifier genes show little overlap, and an evaluation of three reported signatures found poor performance in an external dataset (166). One recent review concluded that an optimal gene signature for prediction of chemoradiotherapy in rectal cancer patients has not yet been found (167). A small number of studies have used transcriptomic data to generate models of 5-FU-based chemotherapy benefit in patients with advanced CRC (168-171). These studies are limited by small numbers of patients and a lack of validation in large patient cohorts. In general, gene signatures developed for predicting risk of tumor recurrence following surgical resection of the primary tumor have not been shown to exhibit predictive value for 5-FU based adjuvant chemotherapy benefit, although in two studies benefit was suggested to be limited to the poor prognosis groups (148,149). The application of transcriptome approaches to targeted biological therapies is an emerging field (172-176). Several of the CRC classification schemes have been suggested to have predictive value for 5-FU-based chemotherapy or radiotherapy, but with apparently conflicting results (Table 1). For example, the mesenchymal “stem cell-like” class of Sadanandam et al. (88) was found to be sensitive to FOLFIRI and radiotherapy, while the mesenchymal “C-type” class of Roepman was associated with 5FU-resistance (91).
The CRC methylome
DNA methylation of cytosines in the context of CpG dinucleotides is a central mechanism of epigenetic control, with essential roles in the maintenance of genome integrity, genomic imprinting, transcriptional regulation, and developmental processes. Multiple approaches for genome-wide studies of DNA methylation patterns have been developed, generally combining DNA analysis by microarrays or NGS with one of three techniques to convert DNA methylation patterns into DNA sequence information or library enrichment: endonuclease digestion, affinity enrichment and bisulphite conversion (177,178).
Genome-wide methylome analyses have highlighted extensive disruption of DNA methylation in CRC. Tumors are typically characterized by global loss of methylation (hypomethylation), predominantly in repetitive sequences, and focal gain in methylation (hypermethylation) in CpG islands, the latter often occurring simultaneously within defined megabase regions (179-181). Hypermethylation within CpG islands is associated with transcriptional silencing of tumor suppressor genes, whilst hypomethylation within gene bodies can affect transcriptional elongation or alternative promoter usage and cause aberrant transcription of oncogenes (182-196). Global loss of methylation may trigger cancer genomic instability and activation of transposons and genes within regions of repetitive sequence (186-188). Both hypo- and hypermethylation occur early in tumorigenesis (189-196), and the average CRC genome carries thousands of methylation changes with marked impact on the cellular transcriptional program (197,198).
Studies have identified a subset of CRCs that exhibit particularly widespread promoter hypermethylation, referred to as the CpG island methylator phenotype (CIMP) (199,200). CIMP is observed in ~30% of CRCs, and presence and extent of CIMP have been used to classify CRC into three major subgroups, CIMP high (CIMP-H), CIMP low (CIMP-L) and non-CIMP (CIMP-0), with distinct clinical and molecular features (201,202). CIMP-H is associated with proximal tumor location, female gender, BRAFV600E mutation, MLH1 methylation and MSI; CIMP-L is characterized by proximal tumor location and KRAS mutation, while CIMP-0 is associated with distal tumor location, TP53 mutation and CIN (202-205).
Aberrant DNA methylation patterns are attractive tumor biomarkers because of their high frequency in neoplasms, and the detection of methylation in DNA isolated from stool and/or blood has emerged as a promising approach for early diagnosis and surveillance of CRC (206,207). Microarray based studies of hypermethylated CpG sites in CRC and benign adenomas have revealed a large number of tumor-specific candidate detection markers (195,208-210). Translation of these candidates into blood- or stool-based diagnostic tests is actively being pursued by academia and industry, involving method development, validation of specificity against normal tissues and other pathologies, and evaluation of performance against routine clinical assays (FOBT, CEA). A recent study evaluating circulating DNA detection of HLTF and HPP1 hypermethylation in addition to CEA serum measurements showed that combination of all three markers outperformed each assay on its own (211). In a related study, Lange et al. suggested that blood-based detection of THBD and C9orf50 hypermethylation outperformed CEA (212). Two diagnostic tests have already been introduced into clinical practice, including a blood-based PCR test for methylated septin-9 (Epi proColon®, ColoVantage® Plasma, RealTime mS9™ kit) (105-109,213-215), and a stool-based test for methylated vimentin (ColoGuard® assay, ColoSure™ assay) (48,110) (Table 2).
The association between CIMP and risk of CRC recurrence has been analyzed extensively, but results remain inconclusive. Several studies indicate CIMP-H as a poor prognostic factor in MSS but not MSI tumors (216,217), while CIMP-L has been suggested to be an indicator of poor outcome regardless of MSI (216,218,219). An association between CIMP and shortened survival was also reported in advanced CRC patients, among whom the contribution of MSI is relatively limited (220). However, there is evidence that the adverse effects associated with CIMP status may be attributable to BRAF mutation (205,221,222). Global hypomethylation as measured by analysis of LINE-1 elements has also been associated with poor outcomes, but data are limited (223,224). Several studies have investigated small numbers of methylated candidate loci not included in CIMP marker panels identifying some evidence for prognostic associations (210,225-228), but no genome-wide methylome studies have been reported.
Epigenetic signatures are increasingly being considered in the context of response to chemotherapeutic and target agents. Recently, Ha et al. correlated genome-wide methylation array data with histopathological rectal tumor regression grade, highlighting hypomethylation of KLHL34 as a candidate predictive marker for sensitivity to preoperative chemoradiation therapy (229). Miyaki et al. related DEX1 hypermethylation and transcriptional silencing, identified by genome-wide methylation sensitive amplified fragment length polymorphism (MS-AFLP) analysis, to resistance of camptothecin 11 (CPT-11) based chemotherapy via inhibition of apoptosis (230). Integrating gene expression microarray analysis and methylation-specific PCR, Tan et al. identified PPP2R2B hypermethylation and transcriptional silencing as a modulator of PDK1-directed Myc signaling and rapamycin sensitivity in CRC (231). CIMP status has been assessed in the context of 5-FU-based adjuvant chemotherapy, but results have not been conclusive. Some investigators have found that 5-FU treatment increases survival in patients with CIMP-H CRC (205,232,233), but others have not replicated this finding (234).
The CRC miRNAome
MicroRNAs (miRNAs) are short (19 to 25 nucleotides), double-stranded, non-protein coding RNAs, that regulate expression of complementary mRNAs at the post-transcriptional level by inducing mRNA degradation or blocking translation into protein. Abnormal miRNA expression profiles are related to clinical and biological behavior of tumors and, given their high stability, have been investigated as robust diagnostic, surveillance, prognostic and predictive biomarkers in cancer tissues and body fluids from cancer patients (235,236). Genomic approaches have mainly utilized qRT-PCR and microarray technologies.
To date, multiple studies have reported unsupervised principle component or cluster analyses of miRNA expression data to classify CRC. Oberg et al. analyzed 315 normal colonic mucosa, tubulovillous adenoma, MSS/proficient mismatch repair (pMMR) sporadic carcinoma, and MSI/deficient mismatch repair (dMMR) sporadic and inherited carcinoma samples using microarrays (237). Unsupervised analysis demonstrated that normal colon tissue, adenomas, MSS/pMMR carcinomas and MSI/dMMR carcinomas were clearly discernible. Consistent with these data, several other studies analyzing MSS/pMMR and MSI/dMMR cancers also found miRNA expression differences between these tumor groups (238-241). One report suggested that Lynch syndrome tumors may display a different miRNA profile as compared to sporadic MSI tumors (242), but this was not noted by Oberg et al. (237). However, overlap between MSI/dMMR associated genes identified across studies is limited. One supervised analysis has suggested miRNA expression differences by CRC location, CIMP, KRAS and TP53 status although this has not been replicated (241). A recent microarray study on 1,141 CRC cases, analyzing 121 miRNAs previously reported with advanced tumor stage and/or survival, verified stage associations for five miRNAs (hsa-miR-145-5p, hsa-miR-31-5p, hsa-miR-200b-3p, hsa-miR-215 and hsa-miR-451a) (243).
miRNA signatures in the blood or stool of CRC patients have been evaluated as an alternative to FOBT testing for CRC diagnosis. Multiple studies have used separate discovery and validation cohorts to derive diagnostic miRNA blood/stool profiles using qRT-PCR panels or microarrays (244-255). Proposed classifiers comprise 1 to 21 miRNAs, with sensitivities of 34-85% and specificities of 68-97% reported across studies. In particular, up-regulation of miRNAs miR21 and miRNA92/miRNA92a have been highlighted in several blood- and stool-based studies for CRC diagnosis (236). To date, signatures have not been validated in independent follow-up reports or been rigorously compared against FOBT testing. Despite these caveats, miRNA signatures show promise as non-invasive CRC biomarkers.
Tumor miRNA signatures have been studied to predict prognosis using qRT-PCR, microarrays and NGS approaches. Several prognostic miRNA signatures for stage I-IV CRC patients have been proposed with little overlap between classifiers (243,256,257). One international study identified a 2-miRNAs classifier for predicting recurrence risk in MSS stage II-III CRC using NGS (258). The most comprehensive discovery and validation study to date has been reported by Zhang et al. (259). Using microarrays, a panel of 35 miRNAs was identified as differentially expressed between 40 paired stage II colon cancer tumors and adjacent normal tissues, and validated in independent samples from 138 patients. Based on these candidate genes, a six-miRNA prognostic classifier (miR-21-5p, miR-20a-5p, miR-103a-3p, miR-106b-5p, miR-143-5p, and miR-215) was built using LASSO Cox regression and validated in an external cohort of 460 stage II patients. Notably, miR-21-5p has been linked to prognosis and advanced tumor stage in multiple other studies (256,260-262). Similar investigations in stage II CRC patients using smaller sample sizes have proposed classifiers with 1-4 miRNAs, and some of these have been validated using additional sample data (239,263,264). None of the proposed prognostic miRNA signatures for stage II CRC overlap. Recently, a study compared the miRNA expression profile in primary cancers and matched liver metastasis using NanoString screening, identifying both primary CRC and serum miRNA signatures with metastasis predictive potential (265).
Multiple studies have investigated the relationship between CRC miRNA signatures and therapy response. A recent microarray-based investigation proposed miRNA-17-5p expression as a predictive marker for 5-FU-based neoadjuvant chemoradiation and adjuvant chemotherapy (266). In metastatic CRC, three studies have identified miRNA signatures associated with the added benefit of oxaliplatin or bevacizumab to 5-FU or capecitabine (267-269), and one study examined the benefit of 5-FU (270). In rectal cancer, miRNA signatures have been proposed to predict response to pre-operative chemoradiotherapy (266,271-275). Interestingly, several studies have suggested that their prognostic classifiers could also predict patients benefit from adjuvant 5-FU based chemotherapy or irinotecan-cetuximab combination therapy (256,257,259,276,277). For example, the 6-miRNA prognostic classifier identified by Zhang et al. also predicted 5-FU treatment response for stage II CRC patients (259,277), while high miR-345 expression identified by Schou et al. was also associated with lack of response for patients to treatment with cetuximab and irinotecan (259,277). Recently, a serum miRNA signature has been proposed as a non-invasive predictor of response to chemotherapy for CRC patients (278).
Conclusions and perspectives
Ongoing global genome characterization efforts are transforming our understanding of CRC biology and pathogenesis. Knowledge of the molecular aberrations driving cancer development—including genome, transcriptome, methylome and miRNAome alterations—can be applied, in principle, to develop integrated approaches for personalized cancer treatment. Recent genome-wide DNA sequencing and copy number studies in CRC have validated established genetic pathways of tumorigenesis and mutator phenotypes, while highlighting extensive mutational heterogeneity and identifying novel cancer gene candidates. Gene expression and DNA methylation data have demonstrated widespread deregulation of the CRC epigenome and indicate the importance of the cell-of-origin, retention of differentiation hierarchy and tumor stroma for CRC molecular classification (87,92,279,280).
Advances in genomics have begun to contribute new tools for clinical diagnosis and management of CRC. Blood- and stool-based tumor DNA sequencing, miRNA detection and DNA methylation assays are being developed for improved population screening, to facilitate surveillance of tumor recurrence and for dynamic monitoring of cancer response to therapy (207,281,282). Direct genomic and transcriptomic analyses of patient tumors are being pursued to provide prognostic and predictive information about the course of disease and benefit of treatment, with the current standard of care already involving assessment of KRAS mutation prior to treatment of metastatic CRC with anti-EGFR antibody therapy (283). Germline pharmacogenomic variation, which we did not consider in this review, further has the potential to predict patient treatment tolerance in order to avoid deleterious side effects (284).
Challenges for translation of genomic-based CRC biomarkers include the need for well-defined clinically characterized cohorts and for standardization regarding specimen collection, handling, and storage. Biomarker translation may further be improved through integration with functional genomics approaches to establish mechanistic rather than correlative links with tumor biology (285). Besides inter-tumor molecular heterogeneity, intra-tumor molecular heterogeneity poses a major hurdle to the translation of genomics findings and remains to be fully elucidated. Efforts focusing on molecular profiling of tumor regional heterogeneity and (epi-) genomic variation between metastatic deposits are ongoing. Besides clonal heterogeneity, hierarchical organization and phenotypic plasticity may play clinically important roles and will be subject of future genomic studies (286,287).
The application of genomic approaches, in particular whole exome sequencing, presents issues beyond the assessment of molecular alterations related to the patient’s original presentation of CRC. Given the comprehensive nature of these tests, incidental findings on clinically relevant variants in genes with no relationship to the primary diagnosis may be made. This raises questions as to whether such findings should be reported back to patients, what method of reporting should be used, and when to disclose these results (288-290).
The revolutionary advances in genomic technologies are enabling the possibility of personalized medicine for CRC. Evolving platforms such as NGS and high-density microarrays are starting to bring precision genomic profiling to the clinic at a reasonable cost. Ongoing innovations in existing applications and clinical informatics algorithms, as well as the many emerging technologies, will continue to advance translational cancer genomics and ultimately contribute to improving patient outcomes.
Acknowledgments
Funding: This work was supported by the Ludwig Institute for Cancer Research, an Australian Rotary Health/District 9780 PhD Scholarship to MP and a National Health and Medical Research Council of Australia (NHMRC) R.D. Wright Biomedical Career Development Fellowship to OMS (APP1062226).
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Jian-Bing Fan) for the series “Application of Genomic Technologies in Cancer Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2015.05.02). The series “Application of Genomic Technologies in Cancer Research” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [PubMed]
- Obrand DI, Gordon PH. Incidence and patterns of recurrence following curative resection for colorectal carcinoma. Dis Colon Rectum 1997;40:15-24. [PubMed]
- Manfredi S, Bouvier AM, Lepage C, et al. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg 2006;93:1115-22. [PubMed]
- Labianca R, Nordlinger B, Beretta GD, et al. Early colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2013;24:vi64-72. [PubMed]
- Meyerhardt JA, Mangu PB, Flynn PJ, et al. Follow-up care, surveillance protocol, and secondary prevention measures for survivors of colorectal cancer: American Society of Clinical Oncology clinical practice guideline endorsement. J Clin Oncol 2013;31:4465-70. [PubMed]
- Benson AB 3rd, Bekaii-Saab T, Chan E, et al. Localized colon cancer, version 3.2013: featured updates to the NCCN Guidelines. J Natl Compr Canc Netw 2013;11:519-28. [PubMed]
- Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300. [PubMed]
- Rex DK, Johnson DA, Anderson JC, et al. American College of Gastroenterology Guidelines for Colorectal Cancer Screening 2009 Am J Gastroenterol 2009;104:739-50. [corrected]. [PubMed]
- Qaseem A, Denberg TD, Hopkins RH Jr, et al. Screening for colorectal cancer: a guidance statement from the American College of Physicians. Ann Intern Med 2012;156:378-86. [PubMed]
- Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006;314:268-74. [PubMed]
- Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science 2007;318:1108-13. [PubMed]
- TCGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [PubMed]
- Ried T, Knutzen R, Steinbeck R, et al. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes and Cancer 1996;15:234-45. [PubMed]
- Meijer GA, Hermsen M, Baak J, et al. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridisation. J Clin Pathol 1998;51:901-9. [PubMed]
- Douglas EJ, Fiegler H, Rowan A, et al. Array comparative genomic hybridization analysis of colorectal cancer cell lines and primary carcinomas. Cancer Res 2004;64:4817-25. [PubMed]
- Nakao K, Mehta K, Fridlyand J, et al. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis 2004;25:1345-57. [PubMed]
- Jones AM, Douglas EJ, Halford SE, et al. Array-CGH analysis of microsatellite-stable, near-diploid bowel cancers and comparison with other types of colorectal carcinoma. Oncogene 2005;24:118-29. [PubMed]
- Sawada T, Sawada E, Yamamoto H, et al. Association between genomic alterations and metastatic behavior of colorectal cancer identified by array-based comparative genomic hybridization. Genes chromosomes cancer 2013;52:140-9. [PubMed]
- Bass AJ, Lawrence MS, Brace LE, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 2011;43:964-8. [PubMed]
- Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012;488:660-4. [PubMed]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382-4. [PubMed]
- Yu J, Wu WK, Liang Q, et al. Disruption of NCOA2 by recurrent fusion with LACTB2 in colorectal cancer. Oncogene 2015; [Epub ahead of print]. [PubMed]
- Timmermann B, Kerick M, Roehr C, et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One 2010;5:e15661 [PubMed]
- Smith CG, Naven M, Harris R, et al. Exome resequencing identifies potential tumor-suppressor genes that predispose to colorectal cancer. Hum mutat 2013;34:1026-34. [PubMed]
- Zhou D, Yang L, Zheng L, et al. Exome capture sequencing of adenoma reveals genetic alterations in multiple cellular pathways at the early stage of colorectal tumorigenesis. PLoS One 2013;8:e53310 [PubMed]
- Gylfe AE, Kondelin J, Turunen M, et al. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology 2013;145:540-3. [PubMed]
- Han SW, Kim HP, Shin JY, et al. Targeted sequencing of cancer-related genes in colorectal cancer using next-generation sequencing. PLoS One 2013;8:e64271 [PubMed]
- Cajuso T, Hänninen UA, Kondelin J, et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int J Cancer 2014;135:611-23. [PubMed]
- Shanmugam V, Ramanathan RK, Lavender NA, et al. Whole genome sequencing reveals potential targets for therapy in patients with refractory KRAS mutated metastatic colorectal cancer. BMC Med Genomics 2014;7:36. [PubMed]
- Brannon AR, Vakiani E, Sylvester BE, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 2014;15:454. [PubMed]
- Vignot S, Lefebvre C, Frampton GM, et al. Comparative analysis of primary tumour and matched metastases in colorectal cancer patients: Evaluation of concordance between genomic and transcriptional profiles. Eur J Cancer 2015;51:791-9. [PubMed]
- Tan IB, Malik S, Ramnarayanan K, et al. High-depth sequencing of over 750 genes supports linear progression of primary tumors and metastases in most patients with liver-limited metastatic colorectal cancer. Genome Biol 2015;16:32. [PubMed]
- Kogita A, Yoshioka Y, Sakai K, et al. Inter- and intra-tumor profiling of multi-regional colon cancer and metastasis. Biochem Biophys Res Commun 2015;458:52-6. [PubMed]
- Dyrsø T, Li J, Wang K, et al. Identification of chromosome aberrations in sporadic microsatellite stable and unstable colorectal cancers using array comparative genomic hybridization. Cancer Genet 2011;204:84-95. [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [PubMed]
- Leary RJ, Kinde I, Diehl F, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med 2010;2:20ra14 [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
- Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430-5. [PubMed]
- Reinert T, Schøler LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2015; [Epub ahead of print]. [PubMed]
- Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med 2008;149:441-50, W81.
- Onouchi S, Matsushita H, Moriya Y, et al. New method for colorectal cancer diagnosis based on SSCP analysis of DNA from exfoliated colonocytes in naturally evacuated feces. Anticancer Res 2008;28:145-50. [PubMed]
- Zou H, Taylor WR, Harrington JJ, et al. High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology 2009;136:459-70. [PubMed]
- Ahlquist DA, Zou H, Domanico M, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology 2012;142:248-56; quiz e25-6.
- Deng L, Qi Z, Zou B, et al. Digital detection of multiple minority mutants in stool DNA for noninvasive colorectal cancer diagnosis. Anal Chem 2012;84:5645-52. [PubMed]
- Li BS, Wang XY, Xu AG, et al. High-resolution melting assay (HRMA) is a simple and sensitive stool-based DNA Test for the detection of mutations in colorectal neoplasms. Clin Colorectal Cancer 2012;11:280-90. [PubMed]
- Zhang H, Wang X, Ma Q, et al. Rapid detection of low-abundance K-ras mutation in stools of colorectal cancer patients using chip-based temperature gradient capillary electrophoresis. Lab Invest 2011;91:788-98. [PubMed]
- Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;371:187-8. [PubMed]
- Tagore KS, Lawson MJ, Yucaitis JA, et al. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer 2003;3:47-53. [PubMed]
- Itzkowitz S, Brand R, Jandorf L, et al. A simplified, noninvasive stool DNA test for colorectal cancer detection. Am J Gastroenterol 2008;103:2862-70. [PubMed]
- Kalimutho M, Del Vecchio Blanco G, Cretella M, et al. A simplified, non-invasive fecal-based DNA integrity assay and iFOBT for colorectal cancer detection. Int J Colorectal Dis 2011;26:583-92. [PubMed]
- Zhang J, Yang S, Xie Y, et al. Detection of methylated tissue factor pathway inhibitor 2 and human long DNA in fecal samples of patients with colorectal cancer in China. Cancer Epidemiol 2012;36:73-7. [PubMed]
- Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut 2008;57:941-50. [PubMed]
- Guastadisegni C, Colafranceschi M, Ottini L, et al. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer 2010;46:2788-98. [PubMed]
- Watanabe T, Kobunai T, Yamamoto Y, et al. Chromosomal instability (CIN) phenotype, CIN high or CIN low, predicts survival for colorectal cancer. J Clin Oncol 2012;30:2256-64. [PubMed]
- Mouradov D, Domingo E, Gibbs P, et al. Survival in stage II/III colorectal cancer is independently predicted by chromosomal and microsatellite instability, but not by specific driver mutations. Am J Gastroenterol 2013;108:1785-93. [PubMed]
- Lanza G, Matteuzzi M, Gafa R, et al. Chromosome 18q allelic loss and prognosis in stage II and III colon cancer. Int J Cancer 1998;79:390-5. [PubMed]
- Martínez-López E, Abad A, Font A, et al. Allelic loss on chromosome 18q as a prognostic marker in stage II colorectal cancer. Gastroenterology 1998;114:1180-7. [PubMed]
- De Angelis PM, Stokke T, Beigi M, et al. Prognostic significance of recurrent chromosomal aberrations detected by comparative genomic hybridization in sporadic colorectal cancer. Int J Colorectal Dis 2001;16:38-45. [PubMed]
- Rooney PH, Boonsong A, McKay JA, et al. Colorectal cancer genomics: evidence for multiple genotypes which influence survival. Br J Cancer 2001;85:1492-8. [PubMed]
- Knösel T, Schluns K, Stein U, et al. Genetic imbalances with impact on survival in colorectal cancer patients. Histopathology 2003;43:323-31. [PubMed]
- Diep CB, Kleivi K, Ribeiro FR, et al. The order of genetic events associated with colorectal cancer progression inferred from meta‐analysis of copy number changes. Genes Chromosomes Cancer 2006;45:31-41. [PubMed]
- Al-Mulla F, Behbehani AI, Bitar MS, et al. Genetic profiling of stage I and II colorectal cancer may predict metastatic relapse. Mod Pathol 2006;19:648-58. [PubMed]
- Kim MY, Yim SH, Kwon MS, et al. RRecurrent genomic alterations with impact on survival in colorectal cancer identified by genome-wide array comparative genomic hybridization. Gastroenterology 2006;131:1913-24. [PubMed]
- Liu XP, Kawauchi S, Oga A, et al. Chromosomal aberrations detected by comparative genomic hybridization predict outcome in patients with colorectal carcinoma. Oncol Rep 2007;17:261-7. [PubMed]
- Kurashina K, Yamashita Y, Ueno T, et al. Chromosome copy number analysis in screening for prognosis-related genomic regions in colorectal carcinoma. Cancer Sci 2008;99:1835-40. [PubMed]
- Sheffer M, Bacolod MD, Zuk O, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009;106:7131-6. [PubMed]
- Brosens RP, Belt EJ, Haan JC, et al. Deletion of chromosome 4q predicts outcome in stage II colon cancer patients. Cell Oncol (Dordr) 2011;34:215-23. [PubMed]
- Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer 2005;41:2060-70. [PubMed]
- Kloosterman WP, Hoogstraat M, Paling O, et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol 2011;12:R103. [PubMed]
- Dewhurst SM, McGranahan N, Burrell RA, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014;4:175-85. [PubMed]
- Yu J, Wu WK, Li X, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut 2015;64:636-45. [PubMed]
- Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603-7. [PubMed]
- Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483:570-5. [PubMed]
- Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013;154:1151-61. [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [PubMed]
- Foltran L, De Maglio G, Pella N, et al. Prognostic role of KRAS, NRAS, BRAF and PIK3CA mutations in advanced colorectal cancer. Future Oncol 2015;11:629-40. [PubMed]
- Peeters M, Oliner KS, Parker A, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clinical Cancer Research 2013;19:1902-12. [PubMed]
- Ciardiello F, Normanno N, Maiello E, et al. Clinical activity of FOLFIRI plus cetuximab according to extended gene mutation status by next-generation sequencing: findings from the CAPRI-GOIM trial. Ann Oncol 2014;25:1756-61. [PubMed]
- Spindler KL, Pallisgaard N, Vogelius I, et al. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012;18:1177-85. [PubMed]
- Mohan S, Heitzer E, Ulz P, et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet 2014;10:e1004271 [PubMed]
- Morelli MP, Overman MJ, Dasari A, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 2015;26:731-6. [PubMed]
- Postma C, Koopman M, Buffart TE, et al. DNA copy number profiles of primary tumors as predictors of response to chemotherapy in advanced colorectal cancer. Ann Oncol 2009;20:1048-56. [PubMed]
- Leon LG, Giovannetti E, Smid K, et al. DNA copy number profiles correlate with outcome in colorectal cancer patients treated with fluoropyrimidine/antifolate-based regimens. Curr Drug Metab 2011;12:956-65. [PubMed]
- Chen Z, Liu Z, Deng X, et al. Chromosomal copy number alterations are associated with persistent lymph node metastasis after chemoradiation in locally advanced rectal cancer. Dis Colon Rectum 2012;55:677-85. [PubMed]
- Grade M, Gaedcke J, Wangsa D, et al. Chromosomal copy number changes of locally advanced rectal cancers treated with preoperative chemoradiotherapy. Cancer Genet Cytogenet 2009;193:19-28. [PubMed]
- De Sousa E. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013;19:614-8. [PubMed]
- Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 2013;19:619-25. [PubMed]
- Marisa L, de Reynies A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453 [PubMed]
- Budinska E, Popovici V, Tejpar S, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol 2013;231:63-76. [PubMed]
- Roepman P, Schlicker A, Tabernero J, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer 2014;134:552-62. [PubMed]
- Calon A, Lonardo E, Berenguer-Llergo A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015;47:320-9. [PubMed]
- Isella C, Terrasi A, Bellomo SE, et al. Stromal contribution to the colorectal cancer transcriptome. Nat Genet 2015;47:312-9. [PubMed]
- Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem 2002;48:1647-53. [PubMed]
- Ahmed FE, James SI, Lysle DT, et al. Improved methods for extracting RNA from exfoliated human colonocytes in stool and RT-PCR analysis. Dig Dis Sci 2004;49:1889-98. [PubMed]
- Collado M, Garcia V, Garcia JM, et al. Genomic profiling of circulating plasma RNA for the analysis of cancer. Clin Chem 2007;53:1860-3. [PubMed]
- Han M, Liew CT, Zhang HW, et al. Novel blood-based, five-gene biomarker set for the detection of colorectal cancer. Clin Cancer Res 2008;14:455-60. [PubMed]
- Yajima S, Ishii M, Matsushita H, et al. Expression profiling of fecal colonocytes for RNA-based screening of colorectal cancer. Int J Oncol 2007;31:1029-37. [PubMed]
- Findeisen P, Rockel M, Nees M, et al. Systematic identification and validation of candidate genes for detection of circulating tumor cells in peripheral blood specimens of colorectal cancer patients. Int J Oncol 2008;33:1001-10. [PubMed]
- Bosch LJ, Carvalho B, Fijneman RJ, et al. Molecular tests for colorectal cancer screening. Clin Colorectal Cancer 2011;10:8-23. [PubMed]
- Bustin SA, Murphy J. RNA biomarkers in colorectal cancer. Methods 2013;59:116-25. [PubMed]
- Wu CW, Sung JJ. Colorectal cancer screening: are stool and blood based tests good enough? Chin Clin Oncol 2013;2:8. [PubMed]
- Marshall KW, Mohr S, Khettabi FE, et al. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer 2010;126:1177-86. [PubMed]
- Novak DJ, Liew GJ, Liew CC. GeneNews Limited: bringing the blood transcriptome to personalized medicine. Pharmacogenomics 2012;13:381-5. [PubMed]
- Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011;9:133. [PubMed]
- deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009;55:1337-46. [PubMed]
- Tóth K, Sipos F, Kalmar A, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One 2012;7:e46000 [PubMed]
- Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 2014;60:1183-91. [PubMed]
- Solomon N, Szostak M, Mak W, et al. editors. The principal and performance characteristics of the Abbott RealTime mS9 colorectal cancer assay. ASCO 2010 molecular markers meeting (abstract# 112); 2010.
- Ned RM, Melillo S, Marrone M. Fecal DNA testing for Colorectal Cancer Screening: the ColoSure™ test. PLoS Curr 2011;3:RRN1220 [PubMed]
- Yip KT, Das PK, Suria D, et al. A case-controlled validation study of a blood-based seven-gene biomarker panel for colorectal cancer in Malaysia. J Exp Clin Cancer Res 2010;29:128. [PubMed]
- O'Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol 2010;28:3937-44. [PubMed]
- Clark-Langone KM, Sangli C, Krishnakumar J, et al. Translating tumor biology into personalized treatment planning: analytical performance characteristics of the Oncotype DX Colon Cancer Assay. BMC cancer 2010;10:691. [PubMed]
- Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 2011;29:4611-9. [PubMed]
- Webber EM, Lin JS, Evelyn P. Whitlock. Oncotype DX tumor gene expression profiling in stage II colon cancer. Application: prognostic, risk prediction. PLoS Curr 2010;2. [PubMed]
- Venook AP, Niedzwiecki D, Lopatin M, et al. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol 2013;31:1775-81. [PubMed]
- Yothers G, O'Connell MJ, Lee M, et al. Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol 2013;31:4512-9. [PubMed]
- Srivastava G, Renfro LA, Behrens RJ, et al. Prospective multicenter study of the impact of oncotype DX colon cancer assay results on treatment recommendations in stage II colon cancer patients. Oncologist 2014;19:492-7. [PubMed]
- Reimers MS, Zeestraten EC, Kuppen PJ, et al. Biomarkers in precision therapy in colorectal cancer. Gastroenterol Rep (Oxf) 2013;1:166-83. [PubMed]
- Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol 2011;29:17-24. [PubMed]
- Maak M, Simon I, Nitsche U, et al. Independent validation of a prognostic genomic signature (ColoPrint) for patients with stage II colon cancer. Ann Surg 2013;257:1053-8. [PubMed]
- Kopetz S, Tabernero J, Rosenberg R, et al. Genomic classifier ColoPrint predicts recurrence in stage II colorectal cancer patients more accurately than clinical factors. Oncologist 2015;20:127-33. [PubMed]
- Lenehan P, Boardman L, Fry D, et al. editors. External validation of a tumor derived 5-gene prognostic signature (OncoDefender-CRC) for recurrence (R) of stages I/II colorectal cancer (CRC). Alexandria: American Society Clinical Oncology, 2011.
- Lenehan PF, Boardman LA, Riegert-Johnson D, et al. Generation and external validation of a tumor-derived 5-gene prognostic signature for recurrence of lymph node-negative, invasive colorectal carcinoma. Cancer 2012;118:5234-44. [PubMed]
- Kennedy RD, Bylesjo M, Kerr P, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol 2011;29:4620-6. [PubMed]
- Bertucci F, Salas S, Eysteries S, et al. Gene expression profiling of colon cancer by DNA microarrays and correlation with histoclinical parameters. Oncogene 2004;23:1377-91. [PubMed]
- Wang Y, Jatkoe T, Zhang Y, et al. Gene expression profiles and molecular markers to predict recurrence of Dukes' B colon cancer. J Clin Oncol 2004;22:1564-71. [PubMed]
- Arango D, Laiho P, Kokko A, et al. Gene-expression profiling predicts recurrence in Dukes' C colorectal cancer. Gastroenterology 2005;129:874-84. [PubMed]
- Barrier A, Lemoine A, Boelle PY, et al. Colon cancer prognosis prediction by gene expression profiling. Oncogene 2005;24:6155-64. [PubMed]
- Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol 2005;23:3526-35. [PubMed]
- Barrier A, Boelle PY, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol 2006;24:4685-91. [PubMed]
- Bandres E, Malumbres R, Cubedo E, et al. A gene signature of 8 genes could identify the risk of recurrence and progression in Dukes' B colon cancer patients. Oncol Rep 2007;17:1089-94. [PubMed]
- Lin YH, Friederichs J, Black MA, et al. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin Cancer Res 2007;13:498-507. [PubMed]
- Yamasaki M, Takemasa I, Komori T, et al. The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. Int J Oncol 2007;30:129-38. [PubMed]
- Anjomshoaa A, Lin YH, Black MA, et al. Reduced expression of a gene proliferation signature is associated with enhanced malignancy in colon cancer. Br J Cancer 2008;99:966-73. [PubMed]
- Jiang Y, Casey G, Lavery IC, et al. Development of a clinically feasible molecular assay to predict recurrence of stage II colon cancer. J Mol Diagn 2008;10:346-54. [PubMed]
- Andersen CL, Christensen LL, Thorsen K, et al. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer 2009;100:511-23. [PubMed]
- Jorissen RN, Gibbs P, Christie M, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res 2009;15:7642-51. [PubMed]
- Staub E, Groene J, Heinze M, et al. An expression module of WIPF1-coexpressed genes identifies patients with favorable prognosis in three tumor types. J Mol Med (Berl) 2009;87:633-44. [PubMed]
- Watanabe T, Kobunai T, Sakamoto E, et al. Gene expression signature for recurrence in stage III colorectal cancers. Cancer 2009;115:283-92. [PubMed]
- Hao JM, Chen JZ, Sui HM, et al. A five-gene signature as a potential predictor of metastasis and survival in colorectal cancer. J Pathol 2010;220:475-89. [PubMed]
- Kalady MF, Dejulius K, Church JM, et al. Gene signature is associated with early stage rectal cancer recurrence. J Am Coll Surg 2010;211:187-95. [PubMed]
- Mettu RK, Wan YW, Habermann JK, et al. A 12-gene genomic instability signature predicts clinical outcomes in multiple cancer types. Int J Biol Markers 2010;25:219-28. [PubMed]
- Pillaire MJ, Selves J, Gordien K, et al. A 'DNA replication' signature of progression and negative outcome in colorectal cancer. Oncogene 2010;29:876-87. [PubMed]
- Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 2010;138:958-68. [PubMed]
- Van Laar RK. An online gene expression assay for determining adjuvant therapy eligibility in patients with stage 2 or 3 colon cancer. Br J Cancer 2010;103:1852-7. [PubMed]
- Watanabe T, Kobunai T, Yamamoto Y, et al. Prediction of liver metastasis after colorectal cancer using reverse transcription-polymerase chain reaction analysis of 10 genes. Eur J Cancer 2010;46:2119-26. [PubMed]
- Oh SC, Park YY, Park ES, et al. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut 2012;61:1291-8. [PubMed]
- Shi M, Beauchamp RD, Zhang B. A network-based gene expression signature informs prognosis and treatment for colorectal cancer patients. PLoS One 2012;7:e41292 [PubMed]
- Sveen A, Agesen TH, Nesbakken A, et al. ColoGuidePro: a prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res 2012;18:6001-10. [PubMed]
- Thorsteinsson M, Kirkeby LT, Hansen R, et al. Gene expression profiles in stages II and III colon cancers: application of a 128-gene signature. Int J Colorectal Dis 2012;27:1579-86. [PubMed]
- Bae T, Rho K, Choi JW, et al. Identification of upstream regulators for prognostic expression signature genes in colorectal cancer. BMC Syst Biol 2013;7:86. [PubMed]
- Giráldez MD, Lozano JJ, Cuatrecasas M, et al. Gene-expression signature of tumor recurrence in patients with stage II and III colon cancer treated with 5'fluoruracil-based adjuvant chemotherapy. Int J Cancer 2013;132:1090-7. [PubMed]
- Chang W, Gao X, Han Y, et al. Gene expression profiling-derived immunohistochemistry signature with high prognostic value in colorectal carcinoma. Gut 2014;63:1457-67. [PubMed]
- Wang L, Shen X, Wang Z, et al. A molecular signature for the prediction of recurrence in colorectal cancer. Mol Cancer 2015;14:22. [PubMed]
- Sanz-Pamplona R, Berenguer A, Cordero D, et al. Clinical value of prognosis gene expression signatures in colorectal cancer: a systematic review. PLoS One 2012;7:e48877 [PubMed]
- Reimers MS, Kuppen PJ, Lee M, et al. Validation of the 12-gene colon cancer recurrence score as a predictor of recurrence risk in stage II and III rectal cancer patients. J Natl Cancer Inst 2014;106. [PubMed]
- Ghadimi BM, Grade M, Difilippantonio MJ, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol 2005;23:1826-38. [PubMed]
- Kim IJ, Lim SB, Kang HC, et al. Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Dis Colon Rectum 2007;50:1342-53. [PubMed]
- Rimkus C, Friederichs J, Boulesteix AL, et al. Microarray-based prediction of tumor response to neoadjuvant radiochemotherapy of patients with locally advanced rectal cancer. Clin Gastroenterol Hepatol 2008;6:53-61. [PubMed]
- Daemen A, Gevaert O, De Bie T, et al. Integrating microarray and proteomics data to predict the response on cetuximab in patients with rectal cancer. Pac Symp Biocomput 2008;166-77. [PubMed]
- Garajová I, Slaby O, Svoboda M, et al. Gene expression profiling in prediction of tumor response to neoadjuvant concomitant chemoradiotherapy in patients with locally advanced rectal carcinoma: pilot study. Cas Lek Cesk 2008;147:381-6. [PubMed]
- Liersch T, Grade M, Gaedcke J, et al. Preoperative chemoradiotherapy in locally advanced rectal cancer: correlation of a gene expression-based response signature with recurrence. Cancer Genet Cytogenet 2009;190:57-65. [PubMed]
- Nishioka M, Shimada M, Kurita N, et al. Gene expression profile can predict pathological response to preoperative chemoradiotherapy in rectal cancer. Cancer Genomics Proteomics 2011;8:87-92. [PubMed]
- Brettingham-Moore KH, Duong CP, Greenawalt DM, et al. Pretreatment transcriptional profiling for predicting response to neoadjuvant chemoradiotherapy in rectal adenocarcinoma. Clin Cancer Res 2011;17:3039-47. [PubMed]
- Brettingham-Moore KH, Duong CP, Heriot AG, et al. Using gene expression profiling to predict response and prognosis in gastrointestinal cancers-the promise and the perils. Ann Surg Oncol 2011;18:1484-91. [PubMed]
- Akiyoshi T, Kobunai T, Watanabe T. Predicting the response to preoperative radiation or chemoradiation by a microarray analysis of the gene expression profiles in rectal cancer. Surg Today 2012;42:713-9. [PubMed]
- Del Rio M, Molina F, Bascoul-Mollevi C, et al. Gene expression signature in advanced colorectal cancer patients select drugs and response for the use of leucovorin, fluorouracil, and irinotecan. J Clin Oncol 2007;25:773-80. [PubMed]
- Watanabe T, Kobunai T, Yamamoto Y, et al. Gene expression signature and response to the use of leucovorin, fluorouracil and oxaliplatin in colorectal cancer patients. Clin Transl Oncol 2011;13:419-25. [PubMed]
- Tsuji S, Midorikawa Y, Takahashi T, et al. Potential responders to FOLFOX therapy for colorectal cancer by Random Forests analysis. Br J Cancer 2012;106:126-32. [PubMed]
- Estevez-Garcia P, Rivera F, Molina-Pinelo S, et al. Gene expression profile predictive of response to chemotherapy in metastatic colorectal cancer. Oncotarget 2015;6:6151-9. [PubMed]
- Debucquoy A, Haustermans K, Daemen A, et al. Molecular response to cetuximab and efficacy of preoperative cetuximab-based chemoradiation in rectal cancer. J Clin Oncol 2009;27:2751-7. [PubMed]
- Grimminger PP, Danenberg P, Dellas K, et al. Biomarkers for cetuximab-based neoadjuvant radiochemotherapy in locally advanced rectal cancer. Clin Cancer Res 2011;17:3469-77. [PubMed]
- Watanabe T, Kobunai T, Yamamoto Y, et al. Gene expression of vascular endothelial growth factor A, thymidylate synthase, and tissue inhibitor of metalloproteinase 3 in prediction of response to bevacizumab treatment in colorectal cancer patients. Dis Colon Rectum 2011;54:1026-35. [PubMed]
- Oliveras-Ferraros C, Vazquez-Martin A, Cufi S, et al. Stem cell property epithelial-to-mesenchymal transition is a core transcriptional network for predicting cetuximab (Erbitux) efficacy in KRAS wild-type tumor cells. J Cell Biochem 2011;112:10-29. [PubMed]
- Pentheroudakis G, Kotoula V, Fountzilas E, et al. A study of gene expression markers for predictive significance for bevacizumab benefit in patients with metastatic colon cancer: a translational research study of the Hellenic Cooperative Oncology Group (HeCOG). BMC Cancer 2014;14:111. [PubMed]
- Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol 2010;28:1106-14. [PubMed]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 2010;11:191-203. [PubMed]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983;301:89-92. [PubMed]
- Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 2012;44:40-6. [PubMed]
- Schweiger MR, Hussong M, Rohr C, et al. Genomics and epigenomics of colorectal cancer. Wiley Interdiscip Rev Syst Biol Med 2013;5:205-19. [PubMed]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 2006;6:107-16. [PubMed]
- Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 2010;466:253-7. [PubMed]
- Hitchins MP, Rapkins RW, Kwok CT, et al. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5'UTR. Cancer cell 2011;20:200-13. [PubMed]
- Shenker N, Flanagan JM. Intragenic DNA methylation: implications of this epigenetic mechanism for cancer research. Br J Cancer 2012;106:248-53. [PubMed]
- Eden A, Gaudet F, Waghmare A, et al. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003;300:455. [PubMed]
- Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 2006;66:8462-9468. [PubMed]
- Howard G, Eiges R, Gaudet F, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 2008;27:404-8. [PubMed]
- Oster B, Thorsen K, Lamy P, et al. Identification and validation of highly frequent CpG island hypermethylation in colorectal adenomas and carcinomas. Int J Cancer 2011;129:2855-66. [PubMed]
- Kibriya MG, Raza M, Jasmine F, et al. A genome-wide DNA methylation study in colorectal carcinoma. BMC Med Genomics 2011;4:50. [PubMed]
- Kim YH, Lee HC, Kim SY, et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol 2011;18:2338-47. [PubMed]
- Spisák S, Kalmar A, Galamb O, et al. Genome-wide screening of genes regulated by DNA methylation in colon cancer development. PLoS One 2012;7:e46215 [PubMed]
- Simmer F, Brinkman AB, Assenov Y, et al. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics 2012;7:1355-67. [PubMed]
- Khamas A, Ishikawa T, Mogushi K, et al. Genome-wide screening for methylation-silenced genes in colorectal cancer. Int J Oncol 2012;41:490-6. [PubMed]
- Naumov VA, Generozov EV, Zaharjevskaya NB, et al. Genome-scale analysis of DNA methylation in colorectal cancer using Infinium HumanMethylation450 BeadChips. Epigenetics 2013;8:921-34. [PubMed]
- Hammoud SS, Cairns BR, Jones DA. Epigenetic regulation of colon cancer and intestinal stem cells. Curr Opin Cell Biol 2013;25:177-83. [PubMed]
- Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science 2013;339:1567-70. [PubMed]
- Luo Y, Wong CJ, Kaz AM, et al. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology 2014;147:418-29. [PubMed]
- Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681-6. [PubMed]
- Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology 2007;132:127-38. [PubMed]
- Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A 2007;104:18654-9. [PubMed]
- Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012;22:271-82. [PubMed]
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787-93. [PubMed]
- Ogino S, Odze RD, Kawasaki T, et al. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. Am J Surg Pathol 2006;30:1175-83. [PubMed]
- Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009;58:90-6. [PubMed]
- Azuara D, Rodriguez-Moranta F, de Oca J, et al. Novel methylation panel for the early detection of colorectal tumors in stool DNA. Clin Colorectal Cancer 2010;9:168-76. [PubMed]
- Wang X, Kuang YY, Hu XT. Advances in epigenetic biomarker research in colorectal cancer. World J Gastroenterol 2014;20:4276-87. [PubMed]
- Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007;3:1709-23. [PubMed]
- Mori Y, Olaru AV, Cheng Y, et al. Novel candidate colorectal cancer biomarkers identified by methylation microarray-based scanning. Endocr Relat Cancer 2011;18:465-78. [PubMed]
- Yi JM, Dhir M, Guzzetta AA, et al. DNA methylation biomarker candidates for early detection of colon cancer. Tumour Biol 2012;33:363-72. [PubMed]
- Philipp AB, Stieber P, Nagel D, et al. Prognostic role of methylated free circulating DNA in colorectal cancer. Int J Cancer 2012;131:2308-19. [PubMed]
- Lange CP, Campan M, Hinoue T, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PLoS One 2012;7:e50266 [PubMed]
- Lange CP, Laird PW. Clinical applications of DNA methylation biomarkers in colorectal cancer. Epigenomics 2013;5:105-8. [PubMed]
- Tóth K, Wasserkort R, Sipos F, et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One 2014;9:e115415 [PubMed]
- Molnár B, Toth K, Bartak BK, et al. Plasma methylated septin 9: a colorectal cancer screening marker. Expert Rev Mol Diagn 2015;15:171-84. [PubMed]
- Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 2008;68:8541-6. [PubMed]
- Kim JH, Shin SH, Kwon HJ, et al. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch 2009;455:485-94. [PubMed]
- Dahlin AM, Palmqvist R, Henriksson ML, et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res 2010;16:1845-55. [PubMed]
- Yagi K, Akagi K, Hayashi H, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res 2010;16:21-33. [PubMed]
- Shen L, Catalano PJ, Benson AB 3rd, et al. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 2007;13:6093-8. [PubMed]
- Lee S, Cho NY, Choi M, et al. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int 2008;58:104-13. [PubMed]
- Pai RK, Jayachandran P, Koong AC, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol 2012;36:744-52. [PubMed]
- Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 2008;100:1734-8. [PubMed]
- Benard A, van de Velde CJ, Lessard L, et al. Epigenetic status of LINE-1 predicts clinical outcome in early-stage rectal cancer. Br J Cancer 2013;109:3073-83. [PubMed]
- Grunau C, Brun ME, Rivals I, et al. BAGE hypomethylation, a new epigenetic biomarker for colon cancer detection. Cancer Epidemiol Biomarkers Prev 2008;17:1374-9. [PubMed]
- Krakowczyk L, Strzelczyk JK, Adamek B, et al. Methylation of the MGMT and p16 genes in sporadic colorectal carcinoma and corresponding normal colonic mucosa. Med Sci Monit 2008;14:BR219-25. [PubMed]
- Nilsson TK, Lof-Ohlin ZM, Sun XF. DNA methylation of the p14ARF, RASSF1A and APC1A genes as an independent prognostic factor in colorectal cancer patients. Int J Oncol 2013;42:127-33. [PubMed]
- Heitzer E, Artl M, Filipits M, et al. Differential survival trends of stage II colorectal cancer patients relate to promoter methylation status of PCDH10, SPARC, and UCHL1. Mod Pathol 2014;27:906-15. [PubMed]
- Ha YJ, Kim CW, Roh SA, et al. Epigenetic regulation of KLHL34 predictive of pathologic response to preoperative chemoradiation therapy in rectal cancer patients. Int J Radiat Oncol Biol Phys 2015;91:650-8. [PubMed]
- Miyaki Y, Suzuki K, Koizumi K, et al. Identification of a potent epigenetic biomarker for resistance to camptothecin and poor outcome to irinotecan-based chemotherapy in colon cancer. Int J Oncol 2012;40:217-26. [PubMed]
- Tan J, Lee PL, Li Z, et al. B55beta-associated PP2A complex controls PDK1-directed myc signaling and modulates rapamycin sensitivity in colorectal cancer. Cancer Cell 2010;18:459-71. [PubMed]
- Van Rijnsoever M, Elsaleh H, Joseph D, et al. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res 2003;9:2898-903. [PubMed]
- Donada M, Bonin S, Barbazza R, et al. Management of stage II colon cancer - the use of molecular biomarkers for adjuvant therapy decision. BMC Gastroenterol 2013;13:36. [PubMed]
- Jover R, Nguyen TP, Perez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology 2011;140:1174-81. [PubMed]
- Turchinovich A, Weiz L, Burwinkel B. Extracellular miRNAs: the mystery of their origin and function. Trends Biochem Sci 2012;37:460-5. [PubMed]
- Mazeh H, Mizrahi I, Ilyayev N, et al. The Diagnostic and Prognostic Role of microRNA in Colorectal Cancer - a Comprehensive review. J Cancer 2013;4:281-95. [PubMed]
- Oberg AL, French AJ, Sarver AL, et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One 2011;6:e20465 [PubMed]
- Lanza G, Ferracin M, Gafa R, et al. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol Cancer 2007;6:54. [PubMed]
- Schepeler T, Reinert JT, Ostenfeld MS, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res 2008;68:6416-24. [PubMed]
- Sarver AL, French AJ, Borralho PM, et al. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 2009;9:401. [PubMed]
- Slattery ML, Wolff E, Hoffman MD, et al. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer 2011;50:196-206. [PubMed]
- Balaguer F, Moreira L, Lozano JJ, et al. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin Cancer Res 2011;17:6239-49. [PubMed]
- Slattery ML, Herrick JS, Mullany LE, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer 2015;137:428-38. [PubMed]
- Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 2009;58:1375-81. [PubMed]
- Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127:118-26. [PubMed]
- Hofsli E, Sjursen W, Prestvik WS, et al. Identification of serum microRNA profiles in colon cancer. Br J Cancer 2013;108:1712-9. [PubMed]
- Luo X, Stock C, Burwinkel B, et al. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS ONE 2013;8:e62880 [PubMed]
- Yong FL, Law CW, Wang CW. Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 2013;13:280. [PubMed]
- Ahmed FE, Ahmed NC, Vos PW, et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genomics Proteomics 2013;10:93-113. [PubMed]
- Koga Y, Yamazaki N, Yamamoto Y, et al. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol Biomarkers Prev 2013;22:1844-52. [PubMed]
- Phua LC, Chue XP, Koh PK, et al. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among Asians. Oncol Rep 2014;32:97-104. [PubMed]
- Wu CW, Ng SC, Dong Y, et al. Identification of microRNA-135b in stool as a potential noninvasive biomarker for colorectal cancer and adenoma. Clin Cancer Res 2014;20:2994-3002. [PubMed]
- Yau TO, Wu CW, Dong Y, et al. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br J Cancer 2014;111:1765-71. [PubMed]
- Wang S, Xiang J, Li Z, et al. A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer 2015;136:152-61. [PubMed]
- Ghanbari R, Mosakhani N, Asadi J, et al. Decreased expression of fecal miR-4478 and miR-1295b-3p in early-stage colorectal cancer. Cancer Biomark 2015;15:195-201. [PubMed]
- Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008;299:425-36. [PubMed]
- Ma Y, Zhang P, Wang F, et al. miR-150 as a potential biomarker associated with prognosis and therapeutic outcome in colorectal cancer. Gut 2012;61:1447-53. [PubMed]
- Goossens-Beumer IJ, Derr RS, Buermans HP, et al. MicroRNA classifier and nomogram for metastasis prediction in colon cancer. Cancer Epidemiol Biomarkers Prev 2015;24:187-97. [PubMed]
- Zhang JX, Song W, Chen ZH, et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: a microRNA expression analysis. Lancet Oncol 2013;14:1295-306. [PubMed]
- Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology 2007;72:397-402. [PubMed]
- Schetter AJ, Nguyen GH, Bowman ED, et al. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res 2009;15:5878-87. [PubMed]
- Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet 2010;200:154-60. [PubMed]
- Weissmann-Brenner A, Kushnir M, Lithwick Yanai G, et al. Tumor microRNA-29a expression and the risk of recurrence in stage II colon cancer. Int J Oncol 2012;40:2097-103. [PubMed]
- Christensen LL, Tobiasen H, Holm A, et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int J Cancer 2013;133:67-78. [PubMed]
- Hur K, Toiyama Y, Schetter AJ, et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst 2015;107. [PubMed]
- Fang L, Li H, Wang L, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget 2014;5:2974-87. [PubMed]
- Rasmussen MH, Jensen NF, Tarpgaard LS, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol 2013;7:637-46. [PubMed]
- Boisen MK, Dehlendorff C, Linnemann D, et al. Tissue microRNAs as predictors of outcome in patients with metastatic colorectal cancer treated with first line Capecitabine and Oxaliplatin with or without Bevacizumab. PLoS One 2014;9:e109430 [PubMed]
- Kjersem JB, Ikdahl T, Lingjaerde OC, et al. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol 2014;8:59-67. [PubMed]
- Molina-Pinelo S, Carnero A, Rivera F, et al. MiR-107 and miR-99a-3p predict chemotherapy response in patients with advanced colorectal cancer. BMC Cancer 2014;14:656. [PubMed]
- Drebber U, Lay M, Wedemeyer I, et al. Altered levels of the onco-microRNA 21 and the tumor-supressor microRNAs 143 and 145 in advanced rectal cancer indicate successful neoadjuvant chemoradiotherapy. Int J Oncol 2011;39:409-15. [PubMed]
- Della Vittoria Scarpati G, Falcetta F, Carlomagno C, et al. A specific miRNA signature correlates with complete pathological response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Int J Radiat Oncol Biol Phys 2012;83:1113-9. [PubMed]
- Svoboda M, Sana J, Fabian P, et al. MicroRNA expression profile associated with response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer patients. Radiat Oncol 2012;7:195. [PubMed]
- Hotchi M, Shimada M, Kurita N, et al. microRNA expression is able to predict response to chemoradiotherapy in rectal cancer. Mol Clin Oncol 2013;1:137-42. [PubMed]
- Kheirelseid EA, Miller N, Chang KH, et al. miRNA expressions in rectal cancer as predictors of response to neoadjuvant chemoradiation therapy. Int J Colorectal Dis 2013;28:247-60. [PubMed]
- Nakajima G, Hayashi K, Xi Y, et al. Non-coding MicroRNAs hsa-let-7g and hsa-miR-181b are Associated with Chemoresponse to S-1 in Colon Cancer. Cancer Genomics Proteomics 2006;3:317-24. [PubMed]
- Schou JV, Rossi S, Jensen BV, et al. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS ONE 2014;9:e99886 [PubMed]
- Zhang J, Zhang K, Bi M, et al. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs 2014;25:346-52. [PubMed]
- Buczacki SJ, Zecchini HI, Nicholson AM, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 2013;495:65-9. [PubMed]
- Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer 2014;14:468-80. [PubMed]
- Newton KF, Newman W, Hill J. Review of biomarkers in colorectal cancer. Colorectal Dis 2012;14:3-17. [PubMed]
- Ganepola GAP, Nizin J, Rutledge JR, et al. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J Gastrointest Oncol 2014;6:83-97. [PubMed]
- Soulières D, Greer W, Magliocco AM, et al. KRAS mutation testing in the treatment of metastatic colorectal cancer with anti-EGFR therapies. Curr Oncol 2010;17:S31-40. [PubMed]
- Wheeler HE, Maitland ML, Dolan ME, et al. Cancer pharmacogenomics: strategies and challenges. Nat Rev Genet 2013;14:23-34. [PubMed]
- Boehm JS, Hahn WC. Towards systematic functional characterization of cancer genomes. Nat Rev Genet 2011;12:487-98. [PubMed]
- Sottoriva A, Verhoeff JJ, Borovski T, et al. Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res 2010;70:46-56. [PubMed]
- Dalerba P, Kalisky T, Sahoo D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 2011;29:1120-7. [PubMed]
- Jackson L, Goldsmith L, O'Connor A, et al. Incidental findings in genetic research and clinical diagnostic tests: a systematic review. Am J Med Genet A 2012;158A:3159-67. [PubMed]
- Jarvik GP, Amendola LM, Berg JS, et al. Return of genomic results to research participants: the floor, the ceiling, and the choices in between. Am J Hum Genet 2014;94:818-26. [PubMed]
- Machini K, Douglas J, Braxton A, et al. Genetic counselors' views and experiences with the clinical integration of genome sequencing. J Genet Couns 2014;23:496-505. [PubMed]