
Cancer genome evolution
Introduction
Like any scientific advancement, new generation sequencing technologies have brought both excitement as well as challenges for cancer research. On one hand, the newly discovered, highly dynamic genetic and epigenetic landscapes of the cancer genome have finally explained why it is so hard to identify common gene mutations in clinical samples, as these dynamics are against the key prediction of the current gene mutation theory of cancer. On the other hand, massive amounts of data from these technologies have also generated confusion in the field (1). For example, despite the clinical reality that the majority of cancer cases display high heterogeneity, most basic researchers have focused on identifying commonly shared genetic patterns. This strategy is largely influenced by results generated over decades from various in vitro and in vivo experimental models, despite the fact that many model systems of cancer come with drastically reduced heterogeneity. However, the gap between basic research and clinical reality is rapidly increasing, and this is one of the key rationales for pushing the cancer genome sequencing project and unbiasedly map the cancer genome landscape and identify these common gene mutations once and for all (1,2). Unexpected by many, the cancer genome sequencing project has forcefully denied such rationale by presenting the highly complicated reality to the research community where every cancer is different, and there is no fixed genomic landscape (3,4). To face this daunting challenge, a new conceptual framework is needed that accounts for the abundant genetic/epigenetic diversity observed. This sentiment has also been shared by some leading researchers, who have admitted that it is not enough to simply continue collecting more sequencing data and suggested that a new paradigm is urgently needed to understand cancer in this age of massive quantities of diverse data (5,6). In contrast, others continue promoting the strategy of sequencing more samples. They are convinced that, by sequencing more samples and using more powerful mathematical and bioinformatics models, the mystery of cancer will ultimately be solved.
To compare these conflicted strategies and reconcile different schools of thought, if possible, we need to understand cancer in the framework of cancer evolution. In particular, genetic/epigenetic variation revealed by sequencing needs to be discussed using the evolutionary mechanism of cancer.
Cancer progression represents an evolutionary process due to its multiple levels of variation (genomic, genetic, epigenetic), inheritance of this variation during progression, and the selective advantages resulting from this heterogeneity. Traditionally, cancer evolution is considered to be a stepwise, Darwinian process, where gene mutations accumulate during waves of clonal expansion. Under this framework, each wave is driven by specific gene mutations, which provide a proliferative advantage to the disease. These powerful molecular drivers are necessary for cancer to progress, and it is believed that key drivers are shared among most patients. Stepwise accumulation is understood as the general pattern of cancer evolution, and any diversification that may occur happens during clonal expansion. Like the concept of Darwinian evolution, cancer evolution is believed to be a continuous, traceable process and is similar to natural selection in the wild.
The evolutionary model of clonal expansion is well accepted in the field of cancer research and is supported by patterns of gene mutations within experimental populations, as well as some exceptional cancer cases such as chronic phase chronic myeloid leukemia (CML-CP) (7). Unfortunately, despite wonderful examples in many experimental systems, the cancer gene mutation theory fails when translated to most clinical cases, and so do stepwise evolutionary explanations of how gene mutations cause cancer. In order to solve this paradox, not only do we need to treat cancer as an evolutionary issue, but we must also search for the correct framework of cancer evolution. Key questions in this search include: Why has the cancer evolutionary concept thus far failed to solve the mystery of cancer? What is the true pattern of cancer evolution? Is current cancer genome sequencing telling us something new regarding cancer evolution? What are the roles of genes, epigenes, and genomes in cancer evolution? What is the new conceptual framework of cancer evolution that takes high levels of genetic heterogeneity into account? Finally, could cancer progression offer a special window to study evolutionary theory in general? One emerging holistic framework, the genome theory of cancer evolution, could serve to answer these questions and clarify confusion in the field.
To address these questions under this proposed framework, we will briefly review the concept of the genome and its importance in cancer evolution, in particular, genome defined system inheritance and its ultimate function in cancer evolution. The genome theory of cancer evolution will be discussed by describing the features of the two phases of cancer evolution and how genome and gene mediated heterogeneity drive macro- and micro-cellular evolution respectively. We will also suggest technologies that focus on genome-level heterogeneity, and we will point out the limitations of current methodologies and statistical approaches that are currently implemented to understand cancer. Finally, the potential applications of cancer genome evolution for understanding organismal evolution will be described.
What is cancer evolution, and why is understanding cancer evolution crucial?
In order to understand cancer evolution, we must first briefly review elementary evolutionary concepts. First, there are three key features that define bio-evolution. These are the following: (I) variations exist within the population; (II) these variations are inheritable and passed on among generations; and (III) these variations provide selective advantages in processes such as competition for space, nutrition, and other resources. Over time, the population will be enriched with certain genetic variations, which are responsible for some dominant features. Second, evolution is traditionally considered as a Darwinian, stepwise process, where the accumulation of small advantages over long periods of time lead to big changes, such as the formation and emergence of new systems or species.
Application of organismal evolutionary concepts in understanding cancer is a logical approach. After all, the cancer process fits well with these three key criteria for evolution, and normal and mutated cells do compete with one another for resources and space in order to successfully grow and dominate cellular populations. The concept of studying cancer evolution dates back to the 1970s (8-12). Classical molecular evolutionary study focuses largely on gradual gene-level change over time, and cancer evolution research has followed this same paradigm. It was believed that a few sequential gene mutations are ultimately needed to transform normal, healthy somatic cells into cancer cells. Clonal expansion thus provides opportunities for cell populations to accumulate the gene mutations necessary to present cancer phenotypes. The reasoning behind this is that molecular pathway change through individual genetic or epigenetic alteration would result in increased fitness, and this would drive cancer growth and progression. Under gene mutation theory, cancer is the result of a stepwise accumulation of small changes in its evolutionary process. Thus, the logical approach would be to look for specific gene mutations that drive cancer evolution. In accordance with this logic, the majority of cancer research is focused on the identification of shared genetic aberrations (e.g., universally common chromosomes or key gene mutations), which would in turn serve as potential diagnostic and therapeutic targets to eradicate cancer. It is important to note that molecular geneticists have identified many gene mutations and pathways, giving the impression that molecular approaches alone would solve the mystery of cancer even without the framework of somatic cell evolution.
Surprisingly, as history and current efforts show, however, this simple concept is difficult to apply to the reality of diverse cancer cases. Aside from exceptional cases including CML-CP, this molecular approach as well as evolutionary explanations have been unsuccessful for the majority of cancers (7). Specifically, solid tumors are, by and large, marked by high degrees of intra- and inter-tumor genome heterogeneity at multiple genetic and non-genetic levels (13,14). This was recently confirmed with high-throughput sequencing (15-17). The high degrees of heterogeneity observed coupled with a lack of common driver mutations have posed a challenge to the general strategy of cancer research and even question the stepwise concept of cancer evolution (1,3,18). What is more troubling is that the efforts to identify shared drivers have resulted in massive amounts of varying and even conflicting data, which have generated confusion and frustration in the field, as these results would suggest that individual genes and pathways offer a minimal contribution to the overall cancer patient population, therefore only holding limited clinical value.
On one hand, we know so much about individual gene mutations, pathways, and the molecular basis for all hallmarks of cancer. On the other hand, the massive sum of diverse data does not make sense under the popular gene mutation theory of cancer. To solve this paradox, we need a new, holistic evolutionary framework that accounts for and unifies this diversity. As we will discuss, shifting research focus from a lower gene level to a higher genome system level embraces this observed multi-level heterogeneity at a single cell resolution while accounting for often neglected large-scale alterations.
Why is it crucial to study cancer at the genome level?
Influenced by gene-centric thinking, cancer research has traditionally focused on the identification and characterization of cancer gene mutations (1). The overwhelming heterogeneity illustrated by current cancer genome sequencing has forced researchers to change the strategy by studying the somatic cell evolutionary process, as an individual gene mutation has limited power in understanding the clinical reality. While the field of cancer evolution research is now picking up steam, as reflected by many important publications, most publications that discuss or acknowledge genome evolution are actually only discussing cancer evolution at the gene level. In fact, very few publications have addressed the issue of cancer evolution at the genome level, despite that cancer genome evolution has become a popular term (16,19).
Genome-based study, which takes into account both overall sequence and three-dimensional topology, has been long ignored in cancer research. Part of the reasoning behind this ignorance may be due to confusion, as the common perception of the genome is that it is merely the collection of genes or the complete DNA sequence. The sequencing of all genes in cancer cells (gene mutation and copy number characterization) has been mistakenly considered as genome research. Due to its highly evolving and re-organizing features, there is no fixed cancer genome. Furthermore, the concept of the genome is not simply the two-dimensional order of nucleotides in DNA! In reality, genome topology serves as a higher level of genetic organization, which governs and defines the genetic network structure. Under the genome, the system can be modified (e.g., through genetic mutation, epigenetic change), however, these lower level changes impact the system at a lesser degree than chromosomal change. To illustrate this important concept, it is essential to redefine what the genome is and why the genomic topology is such an important feature of the bio-system.
Under the genome theory, the genetic information can be classified as “parts inheritance”, or instruction of how to make specific proteins from genes and “system inheritance”, or the directions to assemble a given bio-system. The genetic blueprint does not just provide parts inheritance, but more importantly the system inheritance to organize all parts. A given genome reflected as a new karyotype defines new system inheritance. In nuclei, three-dimensional gene interaction is defined by the order of genes along each chromosome and among different chromosomes, which occupy unique positions; this is opposed to genes, which define parts inheritance (20-22). This concept can be made clear with the following analogy. Consider each individual gene as a unit of building material (e.g., lumber, brick). These can be utilized to construct any kind of building yet, depending on their arrangement, the final results will differ drastically (e.g., cabin, skyscraper). Here, the genome serves as the blueprint that determines how the “building materials” (i.e., genes and their encoded products) will come together to form the structure of the genetic and protein networks. Despite similar gene content, simply changing the genomic topology drastically alters the gene interaction relationship. This has been supported by organismal evolution where distinctive karyotypes can separate different species, and in particular, recent studies where karyotypic alterations were shown to influence gene expression profiles, as well as by single cell sequencing of glioma (23,24). In addition, evidence from yeast studies strongly supports that aneuploidy directly affects gene expression, resulting in phenotypic alteration due to the fact that genome alteration can overcome the lost function of an individual gene (25). The relationship between phenotype and karyotype was recently supported by single cell and population based analyses where genome heterogeneity was linked to growth heterogeneity (26). Thus, the consequence of genome-level alteration is new system formation defined by new system inheritance. This is of high importance in understanding tumor growth and progression, as karyotypic change can result in the generation of an aggressive phenotype, and most cancers are driven by genome replacement coupled with high levels of gene mutation and epigenetic alterations (13). To summarize, from an evolutionary perspective, the role of stochastic genome aberrations in cancer is to increase the evolutionary potential of the disease through increased genome system heterogeneity, resulting in the generation of a wide array of phenotypes and maximized odds for survival upon selection.
One of the major contributions of cancer genome sequencing is the confirmation of previous cytogenetic findings, which have demonstrated that genome level alteration (i.e., karyotypic alteration) is a common phenomenon of most cancers. Genome sequencing and cytogenetic analyses of clinical samples have revealed high rates of chromosomal abnormalities. Subcategories of genome chaos (rapid, stochastic chromosome fragmentation and reorganization) including chromothripsis and chromoplexy have been observed in various types of cancer, and chaotic genomes have been detected in the majority of cases of certain cancer types (16,19,22,27). As discussed, these chromosomal aberrations are necessary for cancer progression as they increase tumor population heterogeneity and thus evolutionary potential. Changes of this magnitude explain the relatively small contribution that individual genes and pathways seem to have in the context of genome alteration-mediated cancer evolution. Chromosomal topology alterations can impact the tumor phenotype more than changing individual pathways by gene mutation, providing explanation why there are so many different types of non-clonal chromosomal aberrations (NCCAs) detected in various cancers and other diseases (3,28-30). This also offers the reasoning why in order to accurately study genome evolution, we must focus on the karyotype level.
To further illustrate the importance of the genome (over individual gene mutations), we have recently introduced the evolutionary mechanism of cancer (31,32). This holistic concept takes a large number of diverse factors into account that can contribute to cancer evolution, including genetic, non-genetic, internal and external factors, as long as it serves as a source of stress to the system and particularly if it induces genome instability (3). We equate the evolutionary mechanism of cancer to the sum of all molecular mechanisms, and this consists of three steps: stress-induced genome system instability, resulting heterogeneity at multiple genetic levels, and somatic cell evolution (4).
The genome theory and evolutionary mechanism of cancer can be understood with application of the multiple level adaptive landscape model (3,22,33), which directly illustrates the relationship between genome change (macro-evolution) and gene changes (micro-evolution) (Figure 1). Here, pathway switching within a cell represents micro-cellular evolution, or small adaptations by local landscape change. Genome switching among cells, however, represents huge adaptation across the overall landscape (macro-cellular evolution). Every genome-mediated global landscape can be achieved through large numbers of pathway-mediated local landscapes. This new strategy accounts for not only the fitness landscape (micro-cellular evolution), but the survival landscape as well (macro-cellular evolution). The key to appreciating the contribution of genome rearrangements lies in understanding the two phases of cancer evolution.
The evolutionary mechanism of cancer and the multiple level adaptive/survival landscape model have also addressed an important issue, which is how to integrate the massive epigenetic dynamics observed from most cancers. Yes, gene mutations and gene regulatory aberrations do not work in isolation, but rather serve to complement each other in bypassing growth controls; however, they mainly address the issue of micro-cellular cancer evolution. In fact, current genomic knowledge of the activation-inactivation relationship between driver genes and a combination of other gene mutations, epigenetic silencing, network regulation, copy number variations, and even chromosomal aberrations are mainly explained within the framework of specific gene mutations and pathways. This is the reason why we have focused mainly on the key challenge to the gene mutation theory, as all genetic/epigenetic alterations are still being linked to gene function [more discussion regarding cancer epigenome can be found in the review from this special issue, see Weisenberger and Liang, 2015 (34)]. Recently, a systems biologist has taken action for such integration (33).
It is important to note that the implications of this understanding extend well beyond cancer and provide insight on many common, complex diseases (21,30,35). Recently, a general model of common and complex diseases has been suggested, where the key is diverse causes lead to genome instability (36). Furthermore, fuzzy inheritance is the basis for such high degree of genome instability. Fuzzy inheritance is a newly identified type of inheritance, where the genetic information at the somatic cell level is much less precise than classical genetics predicted. This mechanism required for evolutionary adaptation ensures necessary variation in cancer and also explains why there is an issue of missing heritability (35-37).
What is the pattern of cancer evolution?
Based on the functional separation of gene and genome, it is important to study the pattern of cancer evolution from both gene and genome point of view. By adapting the new concept that stochastic genomic changes represent an index to measure system instability (traditionally thought of as insignificant “noise”), we performed experiments allowing us to watch cancer evolution in action to compare karyotype changes during cellular immortalization, transformation and drug resistance (10). The following are some discoveries from those studies.
Even prior to The Cancer Genome Atlas (TCGA), the pattern of cancer evolution was already demonstrated to be more complicated than Darwinian stepwise evolution alone (1). The two phases of cancer evolution were originally based on karyotypic observations from an immortalization model where a pattern of clonal and non-clonal expansions was detected, and these phases were recently confirmed in breast cancer using single cell genome sequencing (10,17,38). Cancer evolution is a series of genome-mediated system replacements consisting of dynamic cycles of NCCAs and clonal chromosome aberrations (CCAs) occurring within two evolutionary phases. During the stepwise phase, the majority of cells are clonal across generations, and karyotypic diversification is traceable. The punctuated phase is defined by a high frequency of NCCAs and massive genome reorganization, which break multiple system constraints (e.g., genome integrity, tissue architecture). Cancer progression thus consists of both macro-cellular (genome system replacement) and micro-cellular (genome system modification) evolution. In addition, multiple runs of evolution involve totally different pathways or gene signatures. Further, evolution involves the contributions of multiple genetic levels (genome, gene, epigene), however, their influences vary sharply. Gene-level change modifies an existing system, but genome topology change rapidly creates new systems. Recently, the concepts of macro-micro phases of evolution in cancer have received increased support (39). Interestingly, although the two phases of cancer evolution were recently confirmed with single cell genome sequencing, the punctuated phase can be identified at different genetic levels; however, the correct measures must be taken so that the punctuated phase is not an oversight. For example, the stepwise relationship detected at the sequence level can be found during either the stepwise or punctuated phase at the genome level (10,24,40).
Cancer evolution is much more complicated than traditionally accepted stepwise clonal expansion, and this realization has implications in better understanding cancer progression. First, cancer progression consists of many NCCA/CCA cycles. Second, the NCCA/CCA pattern is dependent on cellular stresses. Under high stress, the phase of cancer evolution can be quickly shifted. Third, in a time of crisis, genome chaos can rapidly change the genomic landscape of the cell population to provide cancer the opportunity to increase heterogeneity and evolutionary potential (through swift creation of drastically altered systems), putting the odds of survival back in cancer’s favor (27). Outlier groups resulting from this process that can better serve niches within the evolutionary landscape could then dominate later cancer progression with new features (e.g., aggressive proliferation) (26).
Since the evolutionary pattern is associated with a wide variety of high stresses, including chemotherapeutics, accounting for macro-cellular evolution has relevance in better understanding drug resistance as well (27,41,42). The current primary standard of care for metastatic patients is application of maximum tolerated doses to eliminate as many tumor cells as possible. While there is initial success with this strategy, there is life after death, as surviving resistant subgroups rapidly repopulate the tumor cell population. With the understanding of stress-induced genome chaos, this paradox becomes clear. Regardless of the specific treatment approach applied, high treatment-related stress will eliminate cells while effectively inducing genome fragmentation and reorganization. These surviving cells with altered genome systems can swiftly recoup lost numbers from the treatment and aggressively drive cancer progression (43) (Horne et al., in preparation). This new mechanism demonstrates that cancer drug resistance is an adaptive process rather than an intrinsic property that is selected for by treatment and must be taken into account in the development of treatment regimens and strategies.
What has TCGA project taught us?
The original goal of TCGA was the identification of common driving gene mutations. It was reasoned that if cancer were a common, stepwise evolutionary process, each patient would represent one snapshot of the same, shared process. By sequencing a large number of samples, the hope was that the overall process as well as main contributing factors would ultimately be identified. These results could then be combined to reveal the overall landscape of the cancer genome and precisely determine the pattern of cancer evolution.
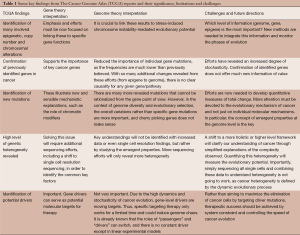
With utilization of technological advancements and large sample sizes, there are waves of excitement that come with releases of major findings and publications from TCGA. As we detail in Table 1, there have certainly been discoveries and achievements from these efforts (the majority of current publications have extensively highlighted most of the achievements of TCGA, and there is no need to repeat them here), but most of these are surprising rather than expected, and they do not fit the original goals of TCGA as rather than finding new key signals in spite of the “noise” of cancer, more heterogeneity is revealed with usage of more powerful technologies. This calls for further evaluation of the limitations as well as challenges of TCGA.
Full table
What methods are needed to study cancer evolution at the genome level?
In order to properly study genome-mediated cancer evolution, focus must be directed at genome-level alterations rather than at other genetic levels. This includes the selection of appropriate techniques to visualize and understand these alterations, as the wrong approach could lead to misinterpretations. For instance, genome chaos cannot be inferred by sequencing alone, as it is a highly rapid and stochastic process rather than traceable and continuous. Importantly, most of the altered genomes observed within cancer samples have already undergone many rounds of genome chaos. Because of this, it is difficult to imagine or infer the process based on end products alone. We have recently demonstrated that the end products and the initial chaotic genomes are drastically different and highly unpredictable (27). However, genome chaos can be precisely followed with in vitro models designed to follow evolution-in-action, and products of genome chaos can be easily defined at the single-cell level with cytogenetic techniques including spectral karyotyping (10,27).
Application of statistical analysis in cancer research warrants reconsideration as well. In light of the strong influence outliers have on the growth and progression of a cancer cell population (26,43), removal of outliers from data sets in the pursuit of “statistically significant” findings has skewed our understanding of cancer. The same goes for average profiling techniques and methodologies, as the average cancer cell likely does not exist within the population (26). Thus, in the effort to identify key cancer “drivers” (i.e., highly expressed markers of averaged results), the actual driving forces of cancer are neglected and eliminated from these analyses (i.e., genome heterogeneity and outlier contributions). To improve the current situation, new analytical platforms are needed that measure heterogeneity and complexity to achieve a true understanding of the disease rather than emphasizing gene and pathway specificity.
As evolution results in different end products with each round (10), it is important for researchers to perform multiple, parallel runs of experiments and incorporate different models in their studies. An individual run may reveal a particular driving factor at a particular time, and a specific linear model may consistently follow a similar molecular progression when repeated. However, these findings only represent conditional possibilities that are quickly muddled when coupled with additional experimental trials or models, or in particular, when compared to clinical data. A general, holistic understanding focused on overall stability and heterogeneity can remedy this confusion. In addition, experiments that follow evolution-in-action will provide greater insight into the disease process than dissecting end products, as the final results will differ with each experiment and thus offer very limited and often misleading information about the overall evolutionary process.
Since cancer is an evolutionary process, and heterogeneity is the key feature of evolution, methods should be developed to monitor the degree of heterogeneity and predict the transition between NCCAs and CCAs. We are currently developing a method to measure the degree of karyotype heterogeneity and complexity. Information from this approach will provide necessary insight for improving patient management (21,36,37,44).
Finally, comparative analysis must be performed to determine the contributions of different genetic levels (epigene, gene, genome) during cancer evolution. Based on previous studies demonstrating the impact of genome-level change on other genetic levels, we anticipate that this type of analysis will undoubtedly show that genome topology alteration drastically alters genetic and epigenetic profiles. We also expect that the role of a particular gene or pathway would change dramatically with karyotypic alteration, as this level of change impacts the entire genetic network. Equally important, quantitative methods are needed to provide improved prediction power in the clinic.
Cancer genome evolution as an ideal model to reveal evolutionary principles
Genome-mediated cancer evolution has offered valuable insight beyond the field of cancer research. For instance, the observed genome/gene dynamics of the evolution-in-action experiments solved the mystery of the main function of sexual reproduction. Traditionally, it is considered that sexual reproduction functions to increase genetic variation. However, under this new paradigm, sex primarily acts to eliminate genomic alterations despite its secondary function of mixing genes (45-47). Thus, sexual reproduction acts as a filter that effectively removes high levels of stochastic genome alterations and maintains species identity.
Cancer genome evolutionary studies have also revealed a trade-off that provides the basis for the many common diseases that lack a clear, causative molecular linkage or heritable factor (30). High-level genome alterations and elevated genome instability have been reported in a wide variety of common diseases including autism, Alzheimer’s disease, Gulf War illness, chronic fatigue syndrome, celiac and Crohn’s disease (36,44,48-52). Interestingly, genome alterations have also been observed in normal, healthy tissues, including the polyploidization of liver cells, skeletal muscle, ovary, placenta, thyroid gland, blood, urothelium, Purkinje neurons, blastocyst mosaicism and trisomy 21 mosaicism in the general population, as well as detected stochastic karyotypic changes caused by environmental and physiological challenges (29,53-57). Whole-genome sequencing of healthy individuals recently revealed an increase of genome-level alteration (58). It is understood that cells at any given time are subject to a wide variety of internal and external stress, under either normal physiological or pathological conditions. Stress, in general, results in many infrequent genome alterations (10,29). Recall that genome-level alterations are more effective at drastically changing the genetic system than gene mutation or epigenetic change. This would suggest that stress-induced genome level change could effectively provide an adaptive advantage for cells against high levels of environmental stress. In addition, genome diversity within normal, healthy tissues allows for complex organ function while providing the genome heterogeneity necessary to account for organ function-associated stress, such as liver-mediated blood detoxification. Thus, stress-induced heterogeneity is necessary for successful adaptation to occur, but the trade-off is potential disease onset (30,36,37,59). If we take into account the new function of sexual reproduction as a filter to eliminate large-scale genome aberrations from the germline, we can understand how system dynamics are promoted for short-term adaptation at the individual level while the accumulation and passing of alterations to offspring is prevented, and this provides clarification behind the “missing heritability” of many common diseases (35).
These studies also led to the realization that macro- and micro-cellular evolution are not simply bridged by time, or in other words, an accumulation of small, stepwise gene-level changes does not often result in large genome topological change over long periods of time. We have analyzed the pattern of cellular evolution by comparing multiple runs of in vitro evolution across several years of culture. Populations that survive always display different genomes, representing macro-cellular evolution (Heng et al., in preparation). In fact, macro- and micro-cellular evolution are two different mechanisms, as evidenced by the genome chaos studies where large-scale genome alteration and new system formation occur within a very short period of time. Micro-cellular evolution, however, acts to tinker and refine the existing system at a much smaller scale (e.g., gene mutation, epigene modification). Interestingly, this distinction between the two mechanisms applies directly to organismal evolution.
The popular framework behind the pattern of organismal evolution needs reconsideration given what we know now about cancer genome evolution. After analyzing a large number of species, King concluded that distinctive karyotypes are the most important features among various species (60). Various groups including our own have promoted this view (46,52,61-63). Thus, rather than a stepwise, Darwinian progression of small changes that lead to speciation events, speciation is likely due to large-scale genome dynamics and preservation of the new species-specific genome through sexual reproduction. Recent genome sequencing of various species supports this idea, as genome alteration is the main event of speciation.
One challenge as well as an opportunity for the field of evolutionary study is to pay more attention to the information derived from cancer progression. Somatic cell evolution provides a unique window to study the interaction of gene mediated micro-cellular evolution and genome replacement mediated macro-cellular evolution. Various in vitro and in vivo systems can serve as platforms to watch evolution-in-action and compare different runs of evolution. Such research opportunities are extremely difficult to access in other systems. Even though cancer differs from many organismal systems, cancer still represents a biological system. This means that cancer should still follow the laws of evolution. Since cancer evolution study has revealed two phases of evolution and can connect the dots between sexual and asexual reproduction and between micro- and macroevolution, the messages derived from cancer evolution extend far beyond somatic cells and are applicable and essential to understanding organismal evolution. One urgent task is to quantitatively study the multiple levels of genetic heterogeneity and how fuzzy inheritance contributes to this heterogeneity (35,36). Furthermore, it is crucial that we understand the similarities and differences that separate cancer genome evolution and organismal genome evolution. The time is now to shift our focus, efforts and technologies onto a new, promising direction and take the next step.
Acknowledgments
This article is part of a series of studies entitled “The mechanisms of somatic cell and organismal evolution”. We thank the reviewers for their constructive suggestions.
Funding: This work was supported by the WSU Office of the Vice President for Research Bridge Funding Grant to HH Heng.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Jian-Bing Fan) for the series “Application of Genomic Technologies in Cancer Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2015.06.01). The series “Application of Genomic Technologies in Cancer Research” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Heng HH. Cancer genome sequencing: the challenges ahead. Bioessays 2007;29:783-94. [PubMed]
- Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am 2007;296:50-7. [PubMed]
- Heng HH, Bremer SW, Stevens JB, et al. Chromosomal instability (CIN): what it is and why it is crucial to cancer evolution. Cancer Metastasis Rev 2013;32:325-40. [PubMed]
- Heng HH, Stevens JB, Bremer SW, et al. Evolutionary mechanisms and diversity in cancer. Adv Cancer Res 2011;112:217-53. [PubMed]
- Weinberg RA. Coming full circle-from endless complexity to simplicity and back again. Cell 2014;157:267-71. [PubMed]
- Roberts NJ, Vogelstein JT, Parmigiani G, et al. The predictive capacity of personal genome sequencing. Sci Transl Med 2012;4:133ra58.
- Horne SD, Stevens JB, Abdallah BY, et al. Why imatinib remains an exception of cancer research. J Cell Physiol 2013;228:665-70. [PubMed]
- Gatenby RA, Silva AS, Gillies RJ, et al. Adaptive therapy. Cancer Res 2009;69:4894-903. [PubMed]
- Greaves M, Maley CC. Clonal evolution in cancer. Nature 2012;481:306-13. [PubMed]
- Heng HH, Stevens JB, Liu G, et al. Stochastic cancer progression driven by non-clonal chromosome aberrations. J Cell Physiol 2006;208:461-72. [PubMed]
- Merlo LM, Pepper JW, Reid BJ, et al. Cancer as an evolutionary and ecological process. Nat Rev Cancer 2006;6:924-35. [PubMed]
- Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23-8. [PubMed]
- Heng HH, Bremer SW, Stevens JB, et al. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol 2009;220:538-47. [PubMed]
- Heppner GH. Tumor heterogeneity. Cancer Res 1984;44:2259-65. [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [PubMed]
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77. [PubMed]
- Wang Y, Waters J, Leung ML, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature 2014;512:155-60. [PubMed]
- Podlaha O, Riester M, De S, et al. Evolution of the cancer genome. Trends Genet 2012;28:155-63. [PubMed]
- Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27-40. [PubMed]
- Heng HH. The genome-centric concept: resynthesis of evolutionary theory. Bioessays 2009;31:512-25. [PubMed]
- Heng HH. Bio-complexity: challenging reductionism. In: Sturmberg JP, Martin CM, editors. Handbook on Systems and Complexity in Health. New York: Springer 2013:193-208.
- Heng HH, Liu G, Stevens JB, et al. Decoding the genome beyond sequencing: the new phase of genomic research. Genomics 2011;98:242-52. [PubMed]
- Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396-401. [PubMed]
- Stevens JB, Liu G, Abdallah BY, et al. Unstable genomes elevate transcriptome dynamics. Int J Cancer 2014;134:2074-87. [PubMed]
- Pavelka N, Rancati G, Zhu J, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 2010;468:321-5. [PubMed]
- Abdallah BY, Horne SD, Stevens JB, et al. Single cell heterogeneity: Why unstable genomes are incompatible with average profiles. Cell Cycle 2013;12:3640-9. [PubMed]
- Liu G, Stevens JB, Horne SD, et al. Genome chaos: Survival strategy during crisis. Cell Cycle 2014;13:528-37. [PubMed]
- Gisselsson D, Hoglund M. Connecting mitotic instability and chromosome aberrations in cancer--can telomeres bridge the gap? Semin Cancer Biol 2005;15:13-23. [PubMed]
- Heng HH, Stevens JB, Liu G, et al. Imaging genome abnormalities in cancer research. Cell Chromosome 2004;3:1. [PubMed]
- Horne SD, Chowdhury SK, Heng HH. Stress, genomic adaptation, and the evolutionary trade-off. Front Genet 2014;5:92. [PubMed]
- Heng HH, Stevens JB, Bremer SW, et al. The evolutionary mechanism of cancer. J Cell Biochem 2010;109:1072-84. [PubMed]
- Ye CJ, Stevens JB, Liu G, et al. Genome based cell population heterogeneity promotes tumorigenicity: the evolutionary mechanism of cancer. J Cell Physiol 2009;219:288-300. [PubMed]
- Huang S. Genetic and non-genetic instability in tumor progression: link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev 2013;32:423-48. [PubMed]
- Weisenberger DJ, Liang G. Contributions of DNA methylation aberrancies in shaping the cancer epigenome. Transl Cancer Res 2015;4:219-34.
- Heng HH. Missing heritability and stochastic genome alterations. Nat Rev Genet 2010;11:813. [PubMed]
- Heng HH, Horne SD, Stevens JB, et al. Heterogeneity mediated system complexity: the ultimate challenge for studying common and complex diseases. Proceedings of the First International Conference on Systems and Complexity in Health. Washington DC, USA. New York: Springer 2015. (In press).
- Heng HH. Debating Cancer: The Paradox in Cancer Research. Singapore: World Scientific Publishing Company, 2015. (In press).
- Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature 2011;472:90-4. [PubMed]
- Klein CA. Selection and adaptation during metastatic cancer progression. Nature 2013;501:365-72. [PubMed]
- Heng HH, Bremer SW, Stevens J, et al. Cancer progression by non-clonal chromosome aberrations. J Cell Biochem 2006;98:1424-35. [PubMed]
- Stevens JB, Abdallah BY, Liu G, et al. Diverse system stresses: common mechanisms of chromosome fragmentation. Cell Death Dis 2011;2:e178 [PubMed]
- Stevens JB, Liu G, Bremer SW, et al. Mitotic cell death by chromosome fragmentation. Cancer Res 2007;67:7686-94. [PubMed]
- Horne SD, Wexler M, Stevens JB, et al. Insights on processes of evolutionary tumor growth. Atlas Genet Cytogenet Oncol Haematol 2015; In press.
- Heng HH, Liu G, Stevens JB, et al. Karyotype heterogeneity and unclassified chromosomal abnormalities. Cytogenet Genome Res 2013;139:144-57. [PubMed]
- Gorelick R, Heng HH. Sex reduces genetic variation: a multidisciplinary review. Evolution 2011;65:1088-98. [PubMed]
- Heng HH. Elimination of altered karyotypes by sexual reproduction preserves species identity. Genome 2007;50:517-24. [PubMed]
- Wilkins AS, Holliday R. The evolution of meiosis from mitosis. Genetics 2009;181:3-12. [PubMed]
- Hojsak I, Gagro A, Petković I, et al. Chromosomal aberrations in peripheral blood lymphocytes in patients with newly diagnosed celiac and Crohn's disease. Eur J Gastroenterol Hepatol 2013;25:22-7. [PubMed]
- Iourov IY, Vorsanova SG, Kurinnaia OS, et al. Molecular karyotyping by array CGH in a Russian cohort of children with intellectual disability, autism, epilepsy and congenital anomalies. Mol Cytogenet 2012;5:46. [PubMed]
- Iourov IY, Vorsanova SG, Yurov YB. Chromosomal mosaicism goes global. Mol Cytogenet 2008;1:26. [PubMed]
- Iourov IY, Vorsanova SG, Yurov YB. Single cell genomics of the brain: focus on neuronal diversity and neuropsychiatric diseases. Curr Genomics 2012;13:477-88. [PubMed]
- Ye CJ, Liu G, Bremer SW, et al. The dynamics of cancer chromosomes and genomes. Cytogenet Genome Res 2007;118:237-46. [PubMed]
- Biesterfeld S, Gerres K, Fischer-Wein G, et al. Polyploidy in non-neoplastic tissues. J Clin Pathol 1994;47:38-42. [PubMed]
- Celton-Morizur S, Desdouets C. Polyploidization of liver cells. Adv Exp Med Biol 2010;676:123-35. [PubMed]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol 2011;27:585-610. [PubMed]
- Fragouli E, Wells D. Aneuploidy in the human blastocyst. Cytogenet Genome Res 2011;133:149-59. [PubMed]
- Hultén MA, Jonasson J, Iwarsson E, et al. Trisomy 21 mosaicism: we may all have a touch of Down syndrome. Cytogenet Genome Res 2013;139:189-92. [PubMed]
- 1000 Genomes Project Consortium, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56-65.
- Horne SD, Pollick SA, Heng HH. Evolutionary mechanism unifies the hallmarks of cancer. Int J Cancer 2015;136:2012-21. [PubMed]
- King M. Species Evolution: The Role of Chromosome Change. New York: Cambridge University Press, 1993.
- Duesberg P, Mandrioli D, McCormack A, et al. Is carcinogenesis a form of speciation? Cell Cycle 2011;10:2100-14. [PubMed]
- Duesberg P, Rasnick D. Aneuploidy, the somatic mutation that makes cancer a species of its own. Cell Motil Cytoskeleton 2000;47:81-107. [PubMed]
- Vincent MD. The animal within: carcinogenesis and the clonal evolution of cancer cells are speciation events sensu stricto. Evolution 2010;64:1173-83. [PubMed]


