
MicroRNAs in head and neck squamous cell carcinoma: a possible challenge as biomarkers, determinants for the choice of therapy and targets for personalized molecular therapies
Introduction
Head and neck cancer are the sixth leading cancer worldwide. About 90% of this cancer is known by squamous cell carcinomas (HNSCC) that emerge from the oral cavity, pharynx, or larynx and for this reason it is a heterogeneous pathology. The major etiologic agents involved in the carcinogenesis of HNSCC are tobacco, alcohol consumption and HPV infection (1-4). The most common cause of mortality in HNSCC patients is high incidence of local recurrences (about 60% of cases) (5,6).
Despite advances in surgery, radiotherapy and chemotherapy, there has been little improvement in HNSCC survival in the last three decades and the clinical outcome has still remained poor. One of the major obstacles in current therapy of HNSCC is radio and chemo-resistance (7,8). Mutation in tumor suppressor genes like TP53 (about 70–80%) and p16INK4a (about 50–80%) are the most frequently detectable genetic alterations reported in HNSCC (9-12). Due to the high rate of genetic and molecular heterogeneity of HNSCC tumors, the diagnosis, prediction, and prognosis of this cancer is particularly challenging (13). An early diagnosis opens the door to future care and treatment but the lack of clinically validated biomarkers for early diagnosis indicates the need for better approaches to identify biomarkers to improve therapeutic efficacy.
miRNAs are one of the best promising biomarkers that are considered as an attractive tool for diagnosis and prognosis of cancer (14-17). They are the class of endogenous expressed small non-coding RNA (19–25 nucleotides) which play a significant role in post transcriptional regulation of genes through interaction with mainly 3’UTR of target mRNAs (18). These small molecules play a crucial role in many biological processes such as cellular development, differentiation, metabolism, proliferation, migration, and apoptosis (19,20).
Several evidences show that aberrant expression of miRNA can lead to cancer initiation, progression and metastasis including HNSCC cancer (18,21). Moreover, they can influence therapy response and clinical outcome in HNSCC (22). Therefore, the significant role of miRNAs cannot be ignored. In this review we will summarize the most recent knowledge and literature about the different implications of microRNAs in HNSCC tumorigenesis and their role to the development of effective targeted therapy.
miRNA biogenesis
The biogenesis of miRNA consists of two pathways. The main pathway for biogenesis and the most studied is “canonical miRNA biogenesis”, the second is called “non-canonical miRNA biogenesis” (23,24) (Figure 1).
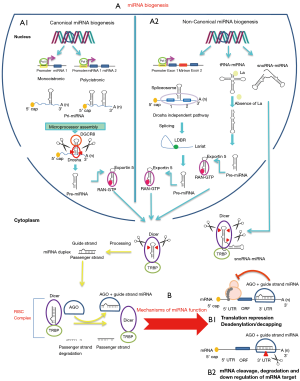
In the canonical miRNA biogenesis, miRNAs are encoded in the genome as a monocistronic or as a cluster which containing several miRNAs. Later these clusters are transcribed to a polycistronic and processed to separate mature miRNAs (23). In most cases, biogenesis starts with transcription by RNA polymerase II (Figure 1, A1 panel). Then the primary transcripts with a stem loop structure (pri-miRNA) are modified by 5’capping, polyadenylation and RNA editing (25). The pri-miRNA is processed in the nucleus by microprocessor complex which containing RNaseIII endonuclease Drosha and the cofactor double-stranded RNA (dsRNA)-binding protein DGCR8 (DiGeorge syndrome critical region gene 8) and cleaved into a smaller precursor termed pre-miRNAs (26). The pre-miR is exported to the cytosol by the exportin 5 (export receptor) in a complex with RAN-GTP through the nuclear pore complex (27). In the cytoplasm the pre-miRNA is cleaved by the endonuclease Dicer and its binding factor TRBP (also known as TRBP2) and produce the miRNA duplex (guide and passenger strand). In the last process called RNA-induced silencing complex (RISC) loading, the miRNA duplex that is produced by Dicer delivered to a core member of AGO family which chooses only one strand as a mature miRNA (guide strand) and interact directly with it. The AGO protein with guide strand separates from the dicer and forms the RISC (23). The other strand of miRNA duplex (passenger strand) is usually released and degraded. However, recent studies show that sometimes passenger strands act as a miRNA and regulate target genes (28).
Besides the canonical biogenesis there are several classes of non- canonical miRNA biogenesis (23,24). Here we overview some of the common non-canonical miRNAs. One of the important classes of these miRNAs which originate from the introns are referred to mirtrons (Figure 1, A2 panel). These miRNAs are transcribed by RNA polymerase II and then spliced. After splicing, the lariat structures are debranched by Lariat debranching enzyme (LDBR) and folded to form pre-miRNA. These miRNAs are processed by Drosha independent, Dicer dependent mechanism. Then they enter the canonical miRNA biogenesis and they are transported to the cytoplasm by exportin 5 (29-32). Some of the non-canonical pre-miRNA derived from tRNA and snoRNA (Figure 1, A2 panel). Despite their expression level, AGO association and mRNA targets are still unclear (33-35).
miRNA function
The specific mechanisms of miRNA function consist of binding seed sequence of miRNA to 3’UTR of the target mRNA in a sequence specific manner. Therefore, they induce inhibition of mRNA translation or degradation and cleavage of mRNA (Figure 1B) (36). Several evidences show that some miRNAs bind not only to the 3’UTR of mRNA but also bind to the CDS (coding sequence) and 5’UTR of target mRNA (37). When miRNAs interact with promoter region can also activate translation or regulate transcription (38,39). Moreover, miRNAs are important for normal animal development (40). In addition, Extracellular miRNAs function as signaling molecules to mediate cell-cell communication (38,41). Chang et al. reported that miR-92a is exported from endothelial cells through extracellular vesicles to macrophages to regulate their functions and enhance atherosclerotic progression (42).
MicroRNAs take part in the tumorigenesis process in HNSCC
By using microRNA expression profiling by microarray analysis, qRT-PCR method, and RNA sequencing, altered miRNA expression in HNSCC cancer has been investigated (1,43,44). miRNAs expression profile is also emerging among the best biomarkers for diagnosis and treatment of cancer, including HNSCC (45). It has been shown that downregulated miRNAs could function as tumor suppressors by negatively regulating oncogenes and overexpressed miRNAs could function as oncogenes by inhibiting tumor suppressor genes (Figure 2) (46,47). In the following sections, we overview some of the recent studies about the function of miRNAs in HNSCC cancer.
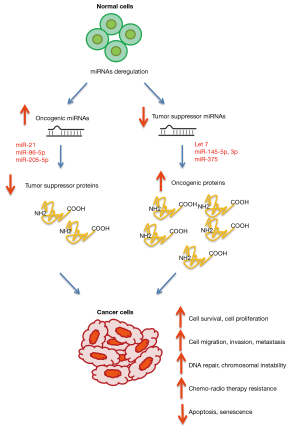
One of the most commonly observed miRNA deregulations in various human cancers including HNSCC, is upregulation of miR-21. The significant correlation between miR-21 overexpression and poor prognosis of HNSCC patients has been described. Therefore, miR-21 could be used as a prognostic biomarker in HNSCC patients (48,49). miR-21 regulates the expression of numerous target genes and these findings support the role of miR-21 as an oncomiR. In nasopharyngeal carcinoma (NPC), overexpression of miR-21, led to increased cell proliferation and suppressed apoptosis through direct targeting PTEN/AKT pathway (50). In oral squamous cell carcinoma, exosomes derived from cisplatin-resistant OSCC cells transferred miR-21 to OSCC parental cells and induced cisplatin resistance by targeting PTEN and PDCD4 (51). Another study observed that sophocarpine (SC) is a chemotherapeutic agent which effectively inhibited proliferation, invasion, and migration of HNSCC cells through downregulation of miR-21 expression by blocking Dicer-mediated miR-21 maturation (52) (Table 1).
Table 1
| miRNAs | Target genes | References |
|---|---|---|
| OncomiRs | ||
| miR-21 | PTEN | (50,51) |
| PDCD4 | (51) | |
| miR-96-5p | PTEN | (53) |
| miR-134 | PDCD7 | (54) |
| miR-155-5p | ARID2 | (55) |
| miR-205-5p | BRCA1, RAD17 | (56) |
| miR-361-3p | OSR2 | (57) |
| miR-501-5p | CLCA4 | (58) |
| miR-762 | PHLPP2,FOXO4 | (59) |
| miR-4295 | NPTX1 | (60) |
| Tumor suppressor miRs | ||
| Let7 family | Ras | (61) |
| Let-7c | IGF1R, HMGA2 | (62) |
| Let-7e-5p | CCR7 | (63) |
| miR-30-5p | EGFR | (64) |
| MET | ||
| IGF1R | ||
| IRS1 | ||
| miR-99a-3p | VEGFA (bioinformatic analysis), STAMBP | (65,66) |
| miR-125a-5p | ERBB2, EIF4EBP1, TXNRD1 | (67) |
| miR-125b | PRXL2A | (68) |
| miR-140-5p | PAK4 | (69) |
| miR-145-3p | MYO1B | (70) |
| miR-145/143 | MDM2 | (71) |
| miR-145-5p | FSCN1 | (72) |
| miR-200 family | QKI | (73) |
| miR-141 | EGFR | (74) |
| miR- 204-5p | Brd4 | (75) |
| JAK2 | (76) | |
| SNAI2, SUZ12, HDAC1, JAK2 | (77) | |
| miR-375 | KLF5 | (78) |
| IGF1R | (79) | |
| JAK2 | (80) | |
| miR-381 | STC2 | (81) |
| miR-625 | SOX4 | (82) |
miRNAs and the relationship with TP53
As mentioned before, in HNSCC, TP53 tumor suppressor gene is the most frequently genetic alteration (about 70–80%) that has been reported (10,83). A group of microRNAs whose expression associates with TP53 mutation was identified (84). Between them, 12 miRNAs whose expression correlates with recurrence-free survival and a group of 4 miRNAs that correlates with cancer-specific survival were determined. Among the 12 miRNA signatures, miR-96-5p emerges as an oncogenic miRNA with prognostic significance in HNSCC. miR-96-5p is one of the members of the cluster miR-96/182/183. It is located on chromosome 7q32.2. Data has shown that overexpression of miR-96-5p was significantly higher in the tumoral tissues in comparison with the non-tumoral tissues (53). This overexpression was significantly stronger in the subset of mutant p53 patients and predicts local recurrence development in HNSCC patients. Furthermore, overexpression of miR-96-5p led to increased cell migration and radio-resistance, chemotherapy resistance in HNSCC cells by directly targeting PTEN and activation of PI3K-AKT signaling pathway (53).
Another study has shown that highly expressed miR-205-5p, in HNSCC expressing mutant p53 proteins, targeted BRCA1 and RAD17 expression leading to inefficient DNA repair and increased chromosomal instability (56). Moreover, this miRNA is among 12 miRNA signatures previously cited and it had the best score in predicting the risk of recurrence in HNSCC (84). Intriguingly, high miR-205-5p expression in HNSCC peritumoral tissues was related to the early detection of minimal residual disease involved in tumor development (56). Surprisingly, in oral squamous cell carcinoma, miR-205-5p has been reported to suppress cancer cell migration and invasion by directly targeted and regulated the tissue inhibitor of met alloproteinases-2 (TIMP-2) gene and act as a tumor suppressor (85). Also, Childs and colleagues have reported that combined under-expression of miR-205 and let-7d showed negative correlation with the survival prognosis of head and neck cancer patients (86). This shows that miRNA expression patterns and biological activity of miRNA are highly specific to different cell-type, tissue context and cellular differentiation status (87).
Recently, it has been shown that long non-coding (lnc) MIR205HG expression was significantly higher in tumoral samples carrying mutant p53 than in tumoral tissues expressing wild-type p53 (88). Moreover, lnc-MIR205HG decreased endogenous miR-590-3p leading to increased cyclin B, cdk1, and YAP protein expression inducing cell proliferation and cell migration (88).
miRNAs as tumor suppressors
In contrast, some miRNAs act as a tumor suppressor by targeting the oncogenes. Several studies have demonstrated that let-7 family is downregulated in HNSCC. Moreover, reports have claimed that let-7 targets a number of genes such as RAS (61), HMGA2 (62) and other oncogenes that is shown in Table 1. As mentioned before, combined under-expression of miR-205 and let-7d were associated with poor survival of HNSCC (86).
Recently, data indicated that let-7e-5p expression was down-regulated in HNSCC tissues and in highly aggressive cells. let-7e-5p inhibited the progression and metastasis of HNSCC by targeting CCR7 (63). Fadhil and colleagues showed that let-7a-5p and miR-3928 were significantly down regulated in saliva of HNSCC patients and salivary let-7a-5p significantly influenced by cancer staging and lymph node metastasis (89).
One of the tumor suppressor miRNAs in HNSCC is miR-145 that targets different genes. miR-143 and miR-145 form a bicistronic cluster in 5q32-q33 (90,91). Accumulating studies showed that both miRNAs are significantly downregulated in cancer tissues (70,92,93). Importantly, Zhang and colleagues identified that miR-143/miR-145 were activated by wild-type p53 consequently, they were involved in the p53 signalling pathway. Moreover, the authors have shown that high levels of miR-143 and miR-145 led to apoptosis of p53 wild-type HNSCCs cells and inhibit both in vitro and in vivo cellular proliferation by reducing MDM2 and enhancing p53 activity (71).
Recently, in laryngeal squamous cell carcinoma (LSCC), the decrease of miR-145-5p increased FSCN1 (fascin actin-bundling protein 1) protein level causing migration, invasion, tumor growth and EMT (epithelial-mesenchymal transition) (72). The authors identified low miR-145-5p/high FSCN1 expressions were associated with poor prognosis and survival in LSCC patients (72). Recent study suggested that carboplatin (analogue of cisplatin) induced the expression of miR-145 in a dose-dependent manner and inhibited the expression of miR-145 direct targets. Furthermore, they showed that pifithrin-α inhibited the p53 in carboplatin-treated cells therefore, reduced miR-145 expression and the miR-145 direct targets (94).
A great number of studies reported that miR-375 act as an antitumor miRNA in HNSCC and this miRNA was strongly repressed in tumor tissues in comparison with normal tissues. Functional analysis showed that miR-375 target several oncogenes that are summarize in Table 1. Moreover, downregulation of miR-375 significantly correlates with cancer survival, poor prognosis, and distant metastasis (95,96). In oral squamous cell carcinoma, low expression level of miR-375 was significantly associated with late stage disease, larger tumor size and increased invasion (97). Furthermore, another study in OSCC revealed the involvement of miR-375 in cancer progression via KLF5 (78). In LSCC miR-375 expression can impede cell proliferation, migration, invasion and enhance apoptosis through IGF1R (79). Also, miR-21/miR-375 ratio is an independent prognostic factor in LSCC patients and this ratio is so specific for disease prediction (98).
Recently, antitumor immunotherapy has great potential to treat several cancers including HNSCC (99). Upregulation of programmed death 1 ligand 1 (PD-L1) in cancer cells and its ligation to PD-1 on T cells promote cancer cell escape from immune surveillance. Secretion of interferon-γ (CD4+ T cell effector cytokine and essential for PD-L1 expression) from natural killer (NK) cells activates the JAK2/STAT1 pathway and increases PD-L1 expression. Wu and colleagues confirmed that miR-375 inhibited IFN-γ-induced PD-L1 surface expression in HNSCC cells via targeting JAK2 (80). Consequently, miR-375 plays a potential role in enhancing T cell-mediated antitumor immune response against HNSCC by inhibiting PD-1/PD-L1 signaling (8075). Table 1 summarizes some of the recent oncogene and tumor suppressor miRNAs in HNSCC (54,55,57-60,64-69,73-77,81,82).
Role of microRNAs in local recurrence and metastatization process in HNSCC
Local recurrences and metastasis are the most common cause of death in HNSCC patients. Therefore, the need to identify biomarkers for early detection of local recurrence and improve survival of HNSCC patients is critical. Migration and invasion are the primary step in the metastatic process. Epithelial cancer cells convert to migrant mesenchymal cells by going through an EMT process and these cells produce new tumors in the same location (recurrence) or different location (metastasis) (100). Therefore, in this process the expression levels of epithelial markers (E-cadherin) are downregulated and the mesenchymal markers (e.g., fibronectin, N-cadherin, vimentin) are upregulated (101). Several transcription factors such as ZEB1, ZEB2, Snail, Slug, and Twist, are involved in the EMT process and the expression levels of these factors are associated with the metastatic status (102). Many recent studies have shown that miRNAs participate in regulating of recurrence, EMT and metastasis (102,103).
Many studies demonstrated that miR-200 family members are downregulated in various cancers including HNSCC and are involved in the EMT process (104-106). The miR-200 family derived from two different chromosome regions: miR-200b/miR-200c/miR-429 cluster which is located on chromosome 1p33.36 and miR-141/miR-200a cluster which is located on 12p13.31 (107). miR-200c has found to negatively regulated the expression of BMI1 and significantly inhibited EMT in malignant HNSCC by reducing the expression of BMI1/ZEB1 (108). In another study, reciprocal correlation between miR-141/mir-200c and ZEB1 was considered. The authors showed that the enforced expression of miR-141 and miR-200c significantly inhibited HNSCC cells migration and led to increased E-cadherin and decreased ZEB1 expression (105). Recent study has suggested that forced expression of miR-200a/b/429 inhibited expression of ZEB1/2 and decreased cell migration in OSCC cell lines by direct targeting QKI (quaking protein). QKI is a member of the signal transduction and activation of RNA (STAR) protein family and it is an important regulator of EMT that has been reported to increase during EMT and modulate EMT-related phenotypes (73).
Downregulation of miR-138 has been frequently reported in HNSCC (109-111). miR-138 showed to regulate the EMT via targeting VIM, ZEB2 and EZH2 genes and control the transcription activity of the E-cadherin in HNSCC. Taken together, this study suggested that miR-138 controls EMT (112). Reinforcing data has shown that ectopic transfection of miR-138 reduced the expression of both RhoC and ROCK2 in tongue squamous cell carcinoma (TSCC) cells and led to decreased cell migration and invasion (113). Recently, in OSCC has been reported that the expression level of miR-138 is negatively correlated with metastasis. The lower expression of miR-138 is related to the higher rate of metastasis and poorer prognosis of the patients. Therefore, miR-138 is emerging as a promising biomarker and therapeutic target for OSCC with metastatic potential (109). Moreover, circulating miRNAs have been identified to be potential biomarker. In support of this, it was reported the positive correlation between decreased serum miR-138 and worse clinical outcome as well as shorter survival (114). Consequently, serum miR-138 might be a potential biomarker for detection and prognosis prediction of HNSCC (114).
Another interesting miRNA involved in carcinogenesis and tumor progression is miR-155-5p found to be involved in cell proliferation, migration and invasion (55,115,116). In OSCC tissues and HNSCC cell lines, miR-155-5p has found significantly upregulated (55,117). This miRNA can regulate EMT by direct targeting ARID2. Downregulation of miR-155-5p by inhibitor led to increase the expression level of epithelial markers E-cadherin and decrease the expression level of mesenchymal markers vimentin and Snail (55). Furthermore, in another study a significant negative correlation was observed between miR-155-5p and E-cadherin expression suggesting its involvement in EMT in OSCC. High expression of miR-155-5p was associated with poorer disease free survival (115,118). Table 2 summarizes some of the miRNAs that are involved in the EMT process in HNSCC.
Table 2
miRNAs and local recurrence events
HNSCC cancers are characterized by a high incidence of local recurrences (6). Interestingly, a signature of 4 miRNAs (miR-96-5p, miR-21-3p, miR-141-3p and miR-130b-3p) was identified whose high expression in OSCC tumors are able to predict local recurrence development independently from the other clinical and pathological variables (120). The authors showed that inhibition of the four prognostic miRNAs expression in led to decrease proliferation rate by downregulation of cyclin D1 (120). Furthermore, Ganci and colleagues identified 35 miRNAs which are deregulated in both tumor and peritumoral tissues, compared with normal matched samples (121). In particular, 4 miRNAs out of these 35 miRNAs (miR-21-3p, miR-21-5p, miR-96-5p and miR-429) predicted local recurrence independently from prognostic clinical variables (121). Therefore, the power of miRNA signature for prediction of local recurrence could not be ignored.
The impact of microRNAs in radioresistance and chemoresistance in HNSCC
The standard cure for HNSCC management consists of radical surgical removal of the tumor accompanied by the adjuvant radiotherapy which can be administered as monotherapy or with chemotherapy and personalized molecular therapy (122,123). Despite all this effort, half of the treated patients are relapsed and do not respond to subsequent therapeutic treatments (124). Distant metastasis formation is most frequent leading to poor prognosis and life quality (124). Furthermore, second primary cancers in the head and neck region (locoregional recurrence) and in other sites as lungs, gastrointestinal tract occur in about 30–50% of HNSCC patients, particularly in those that are HPV-negative (124).
At present, the principal anticancer drug for advanced HNSCC is cisplatin (CDDP), often coupled with radiotherapy (RT) (125,126). Resistance to chemotherapy and RT is associated with poor prognosis, sometimes there is either lack of tumor response or recurrences occur a few weeks after an initial complete response. In these cases, the outcome of patients is much poorer (127). Therefore, CDDP and RT resistance represent the biggest clinical problem in HNSCC patients showing recurrence and because it affects the probabilities of containment of the tumor and greatly affects patient quality of life.
The biological and molecular mechanisms that are the basis of resistance of the tumors to RT and chemotherapies enclosing alterations pathways of DNA damage response and double strand breaks repair, autophagy, apoptosis, proliferation, and angiogenesis. In the last years, many studies have reported that aberrant regulation of these cellular functions is often due to the dysregulation of the miRNA activities and/or expression. The potential of miRNAs to be employed as biomarkers to predict the post-therapy outcomes of HNSCC patients is very high and intriguing.
Signaling pathways that resulted to affect the resistance to the therapies are EGFR, PI3K/AKT, NF-kB, RAS-MAPK, and TGF-β, as well as JAK-STAT signaling (128).
miRNAs and EGFR pathway
EGFR (epidermal growth factor receptor) is overexpressed in about 90% of HNSCC. High EGFR expression correlates with poor prognosis of HNSCC patients and resistance to conventional RT. Diverse miRNAs that target EGFR transcript are downregulated in HNSCC patient tissues contributing to the development of cancer and the acquisition of the malignant phenotype (128,129). Recently, data have shown EGFR and hepatocyte growth factor receptor (c-MET) are directly downregulated by both miR-1 and miR-206 (130). The authors reported that miR-1 and miR-206 expression levels were significantly reduced in HNSCC cell lines and patient cancer tissues compared with matched non-tumoral tissues, while in parallel EGFR and c-MET were expressed at high levels (130). In human genome, miR-1/miR-133a and miR-206/miR-133b are present as miRNA clusters in 20q13.33 and 6p12.1, respectively. Also, miR-133a and miR-133b expression levels frequently downregulated in HNSCC specimens (130). The restoration of miR-1 and miR-206 expression in HNSCC cell lines determined the downregulation of EGFR and c-MET mRNA and the decrease of some oncogenic activities as proliferation, migration, and invasion, showing that these miRNAs act as tumor suppressors in normal cells of oral-pharynx-larynx mucosa (130). miR-1 and miR-206 have a tumor suppressor role also in other type of cancers such as lung cancer, colon cancer and glioblastoma (131-135).
Recently the miR-30 family has identified as an important regulator of signaling and tumor suppressor in HNSCC patients (64). The authors integrated miRNA and mRNA expression, copy number variation, and DNA methylation data from The Cancer Genome Atlas (TCGA), with a functional genome-wide screen revealing that miR-30 family is commonly repressed in HNSCC and showing the inverse correlation with overexpression of predicted target mRNAs deregulated in HNSCC. Pivotal molecules involved in proliferation, differentiation, adhesion, and invasion were repressed and among them also EGFR and c-MET mRNAs (64). Interestingly the treatment with a miR-30a-5p mimic designed for targeted nanomedicine significantly inhibited HNSCC xenograft tumor growth and inhibited growth receptors EGFR and MET in vivo (64).
Recurrent/metastatic HNSCC positive to EGFR expression are currently treated with platinum-based chemotherapy plus the anti-EGFR monoclonal antibody (mAb) cetuximab (136,137). Interestingly, a recent conspicuous and in-depth meta-analysis study revealed important gene-signature (138) and miRNA-signature clusters associated with the treatment response and progression free survival of the patients (139). In particular, in recurrent/metastatic HNSCC patients in the phase II PANI01 trial, the association of Cluster3 gene-signature, representing predictive genes for anti-EGFR cetuximab treatment with seven selected miRNAs (miR-199a-3p, miR-199b-5p, miR-199a-5p, miR-130b-3p, miR-140-5p, miR-214-3p, miR-34a-5p) were significantly associated with panitumumab efficacy, another anti-EGFR-mAb (137). PANI01 tumor samples were analyzed using functional genomics to explore response predictors to anti-EGFR therapy (137).
miRNAs and PI3K/AKT pathway
The activation of EGFR-signaling can induce the downstream phosphorylation cascade of PI3K (phosphatidylinositol-3-kinase) and RAS (140). The activation of PI3K induces the AKT pathway that control different cellular function cell survival, apoptosis, DNA damage response and EMT (140). At the same time, the induction of RAS stimulates a tyrosine kinase cascade, Raf/MEK/MAPK that phosphorylates Myc, FOS, and Jun, some transcriptional factors that lead to cell proliferation (140).
Many miRNAs are resulted to target key components of PI3K and RAS pathways. Interestingly, with the aim to identify prognostic miRNAs for survival, Wu and colleagues performed in silico analysis from the miRNA expression profiles and clinical information of 481 HNSCC patients by TCGA dataset (141). They highlighted two miR-4652-5p and miR-99a-3p associated signatures, 8 hub gene and significant signaling pathways correlated with calcium, as well as cell proliferation, circadian entrainment, EGFR, PI3K-Akt-mTOR, and P53 signaling pathways for estimating the prognosis of patients with HNSCC.
Let-7 miRNA family regulates the expression of RAS and several other cell-cycle related genes (61,142). Several HNSCC tissues showed a low expression of most miRNAs of the let-7 family (143). Recently, Manikandan and colleagues showed that let-7a, let-7d, let-7f, miR-16, and/or miR-143 were able to target 48 genes at different levels of the PI3K/Akt signaling pathway (144). Furthermore, the oncomiRs, miR-29b, miR-142-3p and miR-203 targeted 11 genes of the P53 signaling pathway. They hypothesized that the miRNA deregulated expression may simultaneously induce the constitutive activation of PI3K/Akt pathway and the suppression of the P53 pathway (144). In particular, the biological functions of miR-21 have been extensively investigated in many kinds of cancers, as well as in HNSCC. Its high expression represents a common feature of pathological cell growth, invasion, and chemotherapy resistance because among its target genes are present critical tumor suppressor such as phosphatase and tensin homolog (PTEN), programmed cell death protein 4 (PDCD4), hypoxia inducible factor-1α (HIF-1α) (48,145-148).
Next generation sequencing analyses of HNSCCs have identified frequent gene alterations including TP53, CDKN2A, CASP8, FAT1, NOTCH1, HRAS, PIK3CA, MLL2, and FBXW7 (149). TP53 is the most frequent target of genetic alterations in human cancers, with a prevalence of 40–80% in HNSCC (http://www-p53.iarc.fr). Whole-exome sequencing analyses reported that 58 TP53 mutations occur in HNSCC with a frequency of 72% (Cancer Genome Atlas). The majority of the published results strongly indicate that TP53 mutations produced p53 proteins with a gain-of-function activity contribute to enhance tumor progression, metastatic potential, and/or drug resistance of HNSCC (150). Previously it has shown that TP53 mutations are associated with miRNA expression alteration in HNSCC (84). Some of the TP53-associated miRNAs predicting clinical outcome was also previously reported to be prognostic in HNSCC, as miR-151a-3p, miR-324-5p and miR-21 (151).
Interestingly, miR-96-5p is associated to TP53 status and its high expression level, also associated with other miRNAs, resulted in prediction of local recurrence independently from other clinical co-variables either in tumors or in histologically tumor-free peritumoral tissue (53,84,120). In particular, it has shown that miR-96-5p in the mutant p53 HNSCC increased cell migration and it is involved in radio- and chemo-therapy resistance, at least in part, by directly targeting PTEN transcript and maintaining high activation levels of PI3K-AKT pathway (53).
After many years of research, several groups have highlighted miR-34 as a key tumor suppressor miRNA whose target genes are involved in drug resistance mechanisms. A great number of studies reported that low levels of circulating miR-34 family members or tumor-specific miR-34 expression are associated with low response to chemotherapy (152-155). Re-expression of miR-34 in cancer cells was found to induce apoptosis, cell cycle arrest or senescence (156-158). miR-34a has found significantly repressed in HNSCC tissues and cell lines (159). Studies with tumor samples from HNSCC patients revealed an inverse relationship between miR-34a and survivin as well as miR-34a and E2F3 transcription factor expression. The restoration of miR-34a levels in HNSCC cell lines strongly decreased E2F3 and survivin levels and inhibited cell proliferation and angiogenesis (159).
miR-34 is emerging as a biomarker that could improve cancer precision medicine (160). To date, miR-34 is the only miRNA to have entered a clinical trial (ClinicalTrials.gov Identifiers: NCT02862145; NCT01829971). The delivery system with the best combination of efficacy, biodistribution and safety was the NOV340 technology consisting of ionizable liposome that forms a particle with a diameter of ~120 nm. This study was designed to investigate the biomarkers, pharmacodynamics, and pharmacokinetics of the liposomal micro-ribonucleic acid-34 (miR-34; MRX34) (161,162). Forty-seven patients including eleven cancer types are enrolled and the safety, maximum tolerated dose (MTD) and clinical activity of MRX34 were assessed (161). MRX34 successfully exhibited antitumor activity in the patients with refractory solid tumors but the clinical trial was withdrawn soon because of five immune related adverse events occurred (162). Furthermore, it was observed the unexpected accumulation of MRX34 in healthy tissues due to extravasation of nanocarriers. However, MRX34 in combination with dexamethasone pretreatment, showed a dose-dependent modulation of relevant target genes with a manageable toxicity profile in most of patients providing important information to improve miRNA-based cancer therapy.
The impact of miRNAs in cachexia
About 80% of patients with advanced cancer treated with radio and chemotherapy, including HNSCC patients, are affected by cachexia (163,164). Cachexia is a complex multifactorial syndrome, impossible to treat with food supplements, characterized by the loss of mass and strength and density of skeletal muscle and by the loss of fat mass, with consequent progressive loss of body weight (165). Moreover, cachexia decreases the response to anticancer therapy, and consequently it is correlated with a poor outcome of the patients (165,166). Several groups focused their attention on the identification of molecular mechanisms that govern cachexia cancer-related to select disease biomarkers and new druggable targets. It is crucial to treat cancer cachexia in order to improve patient’s quality of life to render anti-cancer therapy truly effective (167,168). Recently, miRNAs and other non-coding RNA are found to be involved in the pathogenesis of cancer cachexia (169,170). Animal cachectic models showed the association of miRNAs expression with the different features of cachexia (170,171).
In HNSCC there are still not many correlations between miRNA expression and cachexia as in other tumors (171). However, recently Powrózek and colleagues in a cohort of radiotherapy-treated HNSCC patients observed that low levels of miR-130a were associated to higher risk to be classified as cachectic, compared to those with high levels of miR-130a (172). Previously the cytokine TNFα has shown to be a target of miR-130a in cervical cancer cell lines (173). Accordingly, in HNSCC patients the low levels of miR-130a were associated with high plasma levels of TNFα confirming the key role of inflammation in the onset of cancer-associated cachexia (173). Furthermore, it was evaluated the role of circulating miRNAs in a cohort of advanced HNSCC patients treated with radio-chemotherapy (170). In the sera of patients miR-21 was found highly expressed and modulated by radio-chemotherapy treatment. These preliminary results are in line with other observations showing that microvesicles overexpressing miR-21 from lung and pancreatic cancer cell lines induced apoptosis of skeletal muscle cells by activating TLR7 signaling downstream of JNK (174). miR-21 oncogenic activities include increasing of cell proliferation, migration, invasion, metastasis, and resistance to chemotherapeutic treatments demonstrating to be an important biomarker of poor outcome in several solid and hematological malignancies (147,175).
For a wider and more complete reading regarding the involvement of miRNAs associated with cachexia during muscle and adipose tissue wasting in various types of drug-resistant tumors, it can be referred to specific reviews (170,171).
Extracellular microRNAs as biomarkers of HNSCC diagnosis and prognosis
miRNAs are very stable molecules and their expression can be measured in biofluids including plasma, serum, urine, and saliva. This has given more strength to the role of miRNAs as biomarkers in many processes related to HNSCC disease like radio- and chemotherapy resistance, cancer progression, and metastasis (176,177). In the recent years, powerful technologies and highly sensitive instruments have been developed to detect the presence of non-coding RNA in biofluids (178). Some examples of advanced techniques to detect nucleic acids used in liquid biopsy are beads, emulsions, amplification, and magnetics (BEAMing), Cancer Personalized Profiling by deep Sequencing (Capp-Seq), Safe-Sequencing System (Safe-SeqS), ‘Tagged-amplicon deep Sequencing’ (Tam-Seq), in addition to the next-generation sequencing (NGS) already in use in solid tumors (179,180).
miR-21 represents one of the most studied oncomiRs. Its high expression level and its oncogenic role has been reported in different tumors (181). miR-21 high expression was also reported in HNSCC cancer tissues and it was consistently reported to be higher in the plasma, serum-derived exosomes, and in the whole blood of patients compared to healthy controls (182-186). miR-21 expression has recently been reported to be a valuable tool for early detection of tumor recurrence after surgery or chemoradiotherapy and for monitoring patient outcome (183). A longitudinal study showed that the expression level of miR-21 purified by plasma of HNSCC patients was significantly reduced after two months of treatment in case of no relapse. On the other hand, plasma miR-21 did not show a decrease in treated patients who had developed recurrences during the follow-up period (183).
Unlike miR-21, miR-9 is considered to be a miR “tumor suppressor”, in fact its lower expression in plasma was observed at recurrence or metastasis in nasopharyngeal carcinoma and in metastatic patients compared to patients without distant metastases (187). Human papillomavirus (HPV) is an etiological risk factor for HNSCC carcinogenesis and generally HPV positive HNSCC is more radiosensitive with a better prognosis than HPV negative HNSCC (188). miRNA expression patterns are found to be associated with HPV status in HNSCC patients (189). A group of HPV miRNAs were identified and among them miR-15a/miR-16/miR195/miR-497 family, miR-143/miR-145 and the miR-106-363 clusters appear to have an important role in the pathogenesis of HPV (189).
Recent reports highlighted that HNSCC tumor microenvironment can regulate tumor response to radiation therapy throughout a crosstalk between tumor cells and the tumor microenvironment (190). Accordingly, it has been reported that miR-9 found enriched in HPV positive HNSCC exosome were able to enter into the macrophages inducing M1 macrophage polarization via downregulation of PPARδ, a member of the ligand-activated nuclear receptor superfamily involved in inflammation and innate immunity (191). High miR-9 levels rendered HPV positive HNSCC more sensitive to the radiotherapy and further survival analysis suggested that increased level of miR-9 was positively correlated with improved prognosis (191).
About the therapy monitoring, several miRNAs are reported as biomarker from human HNSCC biofluids. For example, high expression of miR-142-3p, miR-186-5p, miR-195-5p, miR-374b-5p and miR-574-3p in the plasma correlated with worse outcome representing promising markers for prognosis and therapy monitoring in the plasma of HNSCC patients (192).
Interestingly, ten HNSCC- or radiation-associated miRNAs were detected in saliva samples (miR-93, miR-125a, miR-142-3p, miR-200a, miR-203, miR-213, let-7a, let-7b, let-7g and let-7i) (193). Furthermore, among them, miR-93 and miR-200a were significantly higher expressed 12 months post-radiotherapy than at baseline. Therefore, these miRNAs seem to be good candidate as biomarkers for the treatment monitoring post-radiation for HNSCC (193).
Recent study identified some miRNAs regulated in OSCC in comparison to a healthy adjacent tissue. The expression of the identified miRNAs in the circulation of same patients was checked (194). Among 48 differentially expressed miRNAs in tissue, 30 miRNAs were also found in the serum of the same patients. miR-32-5p was significantly found at high levels in tumor compared to healthy tissue and it was shown to be up-regulated in the serum. Therefore, this suggests a validated miRNA signature that can be used as potential non-invasive biomarkers of OSCC (194).
HNSCC, as previously mentioned, is typically characterized by a high rate of local recurrences. Biomarkers are able to early indicate the risk of incidence of relapse. Consequently, many research groups are looking for miRNA signatures that can be predictive. Citron and colleagues, by using NanoString array in a cohort of 44 HNSCC patients, identified a signature of four miRNAs that classified the patients at high- or low-risk of recurrence (195). Targets of this signature were genes related EMT process and in particular belonging to druggable pathways (i.e., SP1 and TGFβ) (195). SP1 and TGFβ inhibitors are already used in the clinical, thus the potential of the discovery of this miRNA signature has the immediate translational relevance.
Therefore, the discovery of miRNAs signature with a strong prediction power could give a great benefit to the HNSCC management not only for the diagnosis or the outcome of the disease but also during eventual clinical trials and the efficacy of the treatments.
Challenges for therapies and future aims
On June of 2020, the Food and Drug Administration approved nivolumab (OPDIVO, Bristol-Myers Squibb Co.) for patients with unresectable advanced, recurrent, or metastatic esophageal squamous cell carcinoma (ESCC) after prior fluoropyrimidine- and platinum-based chemotherapy (ATTRACTION-3 clinical trial: NCT02569242). Nivolumab and pembrolizumab, anti-PD-1 immune checkpoint inhibitors, were approved in 2016 for the treatment of patients with recurrent HNSCC refractory to platinum-based regimens (196,197). Immunotherapy is a novel option to treat HNSCC, designed to enhance the immune response to eliminate cancer cells. Thus, predictive biomarkers are urgently needed to classify the patients that may benefit from immunotherapy. Circulating miRNAs are already shown to have potential as predictive biomarkers for anti PD-1/PD-L1 treatment response in diverse cancer diseases (198,199). Recently, it was reported that let-7 family expression was significantly down-regulated in HNSCC, showing a negative correlation with PD-L1 expression (200). Furthermore, in vivo experiments showed that let-7a/b over-expression potentiated anticancer immunotherapy by CTLA-4 antibody treatment offering a novel potential strategy to enhance immune checkpoint therapy for HNSCC (200). To date, only two clinical trials in patients with recurrent/metastatic head and neck squamous cell carcinoma (HNSCC) are present that study the expression of circulating miRNA following treatment with immunotherapy as marginal aim of the project (ClinicalTrials.gov identifiers: NCT03843515 and NCT03723967) (201,202).
From all the studies that have been cited and from many others that unfortunately has not been possible to comment, it is increasingly evident the involvement of miRNAs in pathways relevant to HNSCC cancerogenesis, metastasis, and resistance to treatment. The plethora of the in vitro and in vivo experiments that are collected by diverse researchers have proven that miRNAs may be useful for the early onset of tumor or the recurrence and for individualized treatment strategies. Although there are a very large number of scientific research articles in the literature, there is a considerable discrepancy on the real application of any miRNA-based biomarkers to the prognosis and treatment of HNSCC. In general, we could reflect that HNSCC is a very heterogeneous disease. Several in vitro data sometimes are weak, probably leading to inconsistent results among the studies. Hence, it is emerging the need to increase the number of prospective studies to give solidity to basic and translational research.
However, the combined use of these various miRNA signatures as biomarkers should provide a more personalized approach to clinical management of HNSCC patients which would ultimately lead to better disease outcomes.
Acknowledgments
Funding: The study was supported by
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Alfons Navarro, Joan Josep Castellano and Marina Díaz-Beyá) for the series “Clinic and Therapeutic Potential of Non-coding RNAs in Cancer” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure forms (available at https://dx.doi.org/10.21037/tcr-20-2530). The series “Clinic and Therapeutic Potential of Non-coding RNAs in Cancer” was commissioned by the editorial office without any funding or sponsorship. GB serves as an unpaid editorial board member of Translational Cancer Research from Aug 2019 to Sep 2021. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ganci F, Sacconi A, Manciocco V, et al. Molecular genetics and biology of head and neck squamous cell carcinoma: implications for diagnosis, prognosis and treatment. Agulnik M (ed). Head and Neck Cancer. Rijeka: IntechOpen, 2012;73-122.
- Bose P, Brocktonand NT, Dort JC. Head and neck cancer: from anatomy to biology. Int J Cancer 2013;133:2013-23. [Crossref] [PubMed]
- Argiris A, Karamouzis MV, Raben D, et al. Head and neck cancer. Lancet 2008;371:1695-709. [Crossref] [PubMed]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9-22. [Crossref] [PubMed]
- Leemans CR, Tiwari R, Nauta JJ, et al. Recurrence at the primary site in head and neck cancer and the significance of neck lymph node metastases as a prognostic factor. Cancer 1994;73:187-90. [Crossref] [PubMed]
- Choi P, Chen C. Genetic Expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer 2005;104:1113-28. [Crossref] [PubMed]
- Dimery IW, Hong WK. Overview of combined modality therapies for head and neck cancer. J Natl Cancer Inst 1993;85:95-111. [Crossref] [PubMed]
- Adelstein DJ, Tanb EH, Lavertu P. Treatment of head and neck cancer: the role of chemotherapy. Crit Rev Oncol Hematol 1996;24:97-116. [Crossref] [PubMed]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2-8. [Crossref] [PubMed]
- Balz V, Scheckenbach K, Gotte K, et al. Is the p53 inactivation frequency in squamous cell carcinomas of the head and neck underestimated? Analysis of p53 exons 2-11 and human papillomavirus 16/18 E6 transcripts in 123 unselected tumor specimens. Cancer Res 2003;63:1188-91. [PubMed]
- Thomas GR, Nadiminti H, Regalado J. Molecular Predictors of Clinical Outcome in Patients with Head and Neck Squamous Cell Carcinoma. Int J Exp Pathol 2005;86:347-63. [Crossref] [PubMed]
- Namazie A, Alavi S, Olopade OI, et al. Cyclin D1 Amplification and P16(Mts1/Cdk4i) Deletion Correlate with Poor Prognosis in Head and Neck Tumors. Laryngoscope 2002;112:472-81. [Crossref] [PubMed]
- Alsahafi E, Begg K, Amelio I, et al. Clinical update on head and neck cancer: molecular biology and ongoing challenges. Cell Death and Disease 2019;10:540. [Crossref] [PubMed]
- Babu JM, Prathibha R, Jijith VS, et al. A miR-centric view of head and neck cancers. Biochim Biophys Acta 2011;1816:67-72. [PubMed]
- Di Rocco G, Baldari S, Toietta G. Exosomes and other extracellular vesicles-mediated microRNA delivery for cancer therapy. Transl Cancer Res 2017;6:S1321-30. [Crossref]
- Huang JT, Wang J, Srivastava V, et al. MicroRNA machinery genes as novel biomarkers for cancer. Front. Oncol 2014;4:113. [Crossref] [PubMed]
- Gebert LF, MacRae IJ. Regulation of micRoRNA function in Animals. Nat Rev Mol Cell Biol 2019;20:21-37. [Crossref] [PubMed]
- Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis 2012;33:1126-33. [Crossref] [PubMed]
- Hwang H-W, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br. J. Cancer 2006;94:776-80. [Crossref] [PubMed]
- Peláez N, Carthew RW. Biological robustness and the role of microRNAs: a network perspective. Curr Top Dev Biol 2012;99:237-55. [Crossref] [PubMed]
- Tang G, Tang X, Mendu V, et al. The art of microRNA: Various strategies leading to gene silencing via an ancient pathway. Biochimica et Biophysica Acta 2008;1779:655-62. [Crossref] [PubMed]
- Denaro N, Merlano MC, Russi EG, et al. Non coding RNAs in Head and Neck Squamous Cell Carcinoma (HNSCC): A Clinical Perspective. Anticancer Res 2014;34:6887-96. [PubMed]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 2019;20:5-20. [Crossref] [PubMed]
- Green VA, Weinberg MS. Small RNA-Induced Transcriptional Gene Regulation in Mammals: Mechanisms, Therapeutic Applications, and Scope Within the Genome. Progress in Molecular Biology and Translational Science 2011;102:11-46. [Crossref] [PubMed]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004;23:4051-60. [Crossref] [PubMed]
- Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003;425:415-9. [Crossref] [PubMed]
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP- dependent dsRNA- binding protein that mediates nuclear export of pre- mi-RNAs. RNA 2004;10:185-91. [Crossref] [PubMed]
- Kobayashi H, Tomari Y. RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta 2016;1859:71-81. [Crossref] [PubMed]
- Berezikov E, Chung WJ, Willis J, et al. Mammalian mirtron genes. Mol Cell 2007;28:328-36. [Crossref] [PubMed]
- Wen J, Ladewig E, Shenker S, et al. Analysis of nearly one thousand mammalian mirtrons reveals novel features of dicer substrates. PLOS Comput Biol 2015;11:e1004441. [Crossref] [PubMed]
- Hasler D, Meister G. From tRNA to miRNA: RNA-folding contributes to correct entry into noncoding RNA pathways. FEBS Lett 2016;590:2354-63. [Crossref] [PubMed]
- Hasler D, Lehman G, Murakawa Y, et al. The lupus autoantigen La prevents mis- channeling of tRNA fragments into the human microRNA pathway. Mol Cell 2016;63:110-24. [Crossref] [PubMed]
- Taft RJ, Glazov EA, Lassmann T, et al. Small RNAs derived from snoRNAs. RNA 2009;15:1233-40. [Crossref] [PubMed]
- Ender C, Krek A, Friedländer M, et al. A human snoRNA with microRNA- like functions. Mol Cell 2008;32:519-28. [Crossref] [PubMed]
- Cifuentes D, Xue H, Taylor DW, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010;328:1694-8. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Zhang K, Zahng X, Cai Z, et al. A novel class of microRNA-recognition elements that function only within open reading frames. Nat Struct Mol Biol 2018;25:1019-27. [Crossref] [PubMed]
- O'Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018;9:402. [Crossref] [PubMed]
- Dharap A, Pokrzywa C, Murali S, et al. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS One 2013;8:e79467. [Crossref] [PubMed]
- Fu G, Brkić J, Hayder H, et al. MicroRNAs in Human Placental Development and Pregnancy Complications. Int J Mol Sci 2013;14:5519-44. [Crossref] [PubMed]
- Kosaka N, Yoshioka Y, Hagiwara K, et al. Trash or Treasure: extracellular microRNAs and cell-to-cell communication. Front Genet 2013;4:173. [Crossref] [PubMed]
- Chang YJ, Li YS, Wu CC, et al. Extracellular MicroRNA-92a Mediates Endothelial Cell-Macrophage Communication. Arterioscler Thromb Vasc Biol 2019;39:2492-504. [Crossref] [PubMed]
- Shiiba M, Uzawa K, Tanzawa H. MicroRNAs in Head and Neck Squamous Cell Carcinoma (HNSCC) and Oral Squamous Cell Carcinoma (OSCC). Cancers 2010;2:653-69. [Crossref] [PubMed]
- Yan L, Zhan C, Wu J, et al. Expression profile analysis of head and neck squamous cell carcinomas using data from The Cancer Genome Atlas. Molecular Medicine Reports 2016;13:4259-65. [Crossref] [PubMed]
- Jamali Z, Asl Aminabadi N, Attaran R, et al. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncology 2015;51:321-31. [Crossref] [PubMed]
- Masood Y, Kqueen CY, Rajadurai P. Role of miRNA in Head and Neck Squamous Cell Carcinoma. Expert Rev Anticancer Ther 2015;15:183-97. [Crossref] [PubMed]
- Blandino G, Fazi F, Donzelli S. Tumor suppressor microRNAs: A novel non-coding alliance against cancer. FEBS Letters 2014;588:2639-52. [Crossref] [PubMed]
- Irimie-Aghiorghiesei AI, Pop-Bica C, Pintea S, et al. Prognostic Value of MiR-21: An Updated Meta-Analysis in Head and Neck Squamous Cell Carcinoma (HNSCC). J Clin Med 2019;8:2041. [Crossref] [PubMed]
- Irani S. miRNAs Signature in Head and Neck Squamous Cell Carcinoma Metastasis: A Literature Review. J Dent (Shiraz) 2016;17:71-83. [PubMed]
- Ou H, Li Y, Kang M. Activation of miR-21 by STAT3 Induces Proliferation and Suppresses Apoptosis in Nasopharyngeal Carcinoma by Targeting PTEN Gene. PLoS One 2014;9:e109929. [Crossref] [PubMed]
- Liu T, Chen G, Sun D, et al. Exosomes Containing miR-21 Transfer the Characteristic of Cisplatin Resistance by Targeting PTEN and PDCD4 in Oral Squamous Cell Carcinoma. Acta Biochim Biophys Sin (Shanghai) 2017;49:808-16. [Crossref] [PubMed]
- Liu W, Zhang B, Chen G, et al. Targeting miR-21 with Sophocarpine Inhibits Tumor Progression and Reverses Epithelial-Mesenchymal Transition in Head and Neck Cancer. Molecular Therapy 2017;25:2129-39. [Crossref] [PubMed]
- Vahabi M, Pulito C, Sacconi A, et al. miR-96-5p targets PTEN expression affecting radio-chemosensitivity of HNSCC cells. J Exp Clin Cancer Res 2019;38:141. [Crossref] [PubMed]
- Peng SY, Tu HF, Yang CC, et al. miR-134 targets PDCD7 to reduce E-cadherin expression and enhance oral cancer progression. Int J Cancer 2018;143:2892-904. [Crossref] [PubMed]
- Wu M, Duan Q, Liu X, et al. MiR-155-5p promotes oral cancer progression by targeting chromatin remodeling gene ARID2. Biomed Pharmacother 2020;122:109696. [Crossref] [PubMed]
- Valenti F, Sacconi A, Ganci F, et al. The miR-205-5p/BRCA1/RAD17 Axis Promotes Genomic Instability in Head and Neck Squamous Cell Carcinomas. Cancers 2019;11:1347. [Crossref] [PubMed]
- Ogawa H, Ogawa KI, Tokuzen N, et al. MicroRNA-361-3p is a potent therapeutic target for oral squamous cell carcinoma. Cancer Sci 2020;111:1645-51. [Crossref] [PubMed]
- Li B, Jiang YP, Zhu J, et al. MiR-501-5p acts as an energetic regulator in head and neck squamous cell carcinoma cells growth and aggressiveness via reducing CLCA4. Mol Biol Rep 2020;47:2181-7. [Crossref] [PubMed]
- Chen S, Zhang JY, Sun LS, et al. miR-762 Promotes Malignant Development of Head and Neck Squamous Cell Carcinoma by Targeting PHLPP2 and FOXO4. Onco Targets Ther 2019;12:11425-11436. [Crossref] [PubMed]
- Lu S, Zhou C, Zou B, et al. MiR-4295 facilitates cell proliferation and metastasis in head and neck squamous cell carcinoma by targeting NPTX1. Genes Immun 2020;21:4-12. [Crossref] [PubMed]
- Johnson SM, Grosshans H, Shingara J. RAS Is Regulated by the let-7 MicroRNA Family. Cell 2005;120:635-47. [Crossref] [PubMed]
- Hou B, Ishinaga H, Midorikawa K, et al. Let-7c inhibits migration and epithelial-mesenchymal transition in head and neck squamous cell carcinoma by targeting IGF1R and HMGA2. Oncotarget 2018;9:8927-40. [Crossref] [PubMed]
- Wang S, Jin S, Liu MD, et al. Hsa-let-7e-5p Inhibits the Proliferation and Metastasis of Head and Neck Squamous Cell Carcinoma Cells by Targeting Chemokine Receptor 7. J Cancer 2019;10:1941-8. [Crossref] [PubMed]
- Saleh AD, Cheng H, Martin SE, et al. Integrated Genomic and Functional microRNA Analysis Identifies miR-30-5p as a Tumor Suppressor and Potential Therapeutic Nanomedicine in Head and Neck Cancer. Clin Cancer Res 2019;25:2860-73. [Crossref] [PubMed]
- Okada R, Koshizuka K, Yamada Y, et al. Regulation of Oncogenic Targets by miR-99a-3p (Passenger Strand of miR-99a-Duplex) in Head and Neck Squamous Cell Carcinoma. Cells 2019;8:1535. [Crossref] [PubMed]
- Wei GG, Guo WP, Tang ZY, et al. Expression level and prospective mechanism of miRNA-99a-3p in head and neck squamous cell carcinoma based on miRNA-chip and miRNA sequencing data in 1, 167 cases. Pathol Res Pract 2019;215:963-976. [Crossref] [PubMed]
- Vo DT, Karanam NK, Ding L, et al. miR-125a-5p Functions as Tumor Suppressor microRNA And Is a Marker of Locoregional Recurrence And Poor prognosis in Head And Neck Cancer. Neoplasia 2019;21:849-62. [Crossref] [PubMed]
- Chen YF, Wei YY, Yang CC, et al. miR-125b suppresses oral oncogenicity by targeting the anti-oxidative gene PRXL2A. Redox Biol 2019;22:101140. [Crossref] [PubMed]
- Peng M, Pang C. MicroRNA‐140‐5p inhibits the tumorigenesis of oral squamous cell carcinoma by targeting p21‐activated kinase 4. Cell Biol Int 2019; [Crossref] [PubMed]
- Yamada Y, Koshizuka K, Hanazawa T, et al. Passenger strand of miR-145-3p acts as a tumor-suppressor by targeting MYO1B in head and neck squamous cell carcinoma. Int J Oncol 2018;52:166-78. [PubMed]
- Zhang J, Sun Q, Zhang Z, et al. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene 2013;32:61-9. [Crossref] [PubMed]
- Gao W, Zhang C, Li W, et al. Promoter Methylation-Regulated miR-145-5p Inhibits Laryngeal Squamous Cell Carcinoma Progression by Targeting FSCN1. Molecular Therapy 2019;27:365-79. [Crossref] [PubMed]
- Kim EJ, Kim JS, Lee S, et al. QKI, a miR-200 target gene, suppresses epithelial-to-mesenchymal transition and tumor growth. Int J Cancer 2019;145:1585-95. [Crossref] [PubMed]
- Zhao Z, Gao D, Ma T, et al. MicroRNA-141 suppresses growth and metastatic potential of head and neck squamous cell carcinoma. Aging (Albany NY) 2019;11:921-32. [Crossref] [PubMed]
- Wang C, Zhang Y, Zhou D, et al. miR 204 enhances p27 mRNA stability by targeting Brd4 in head and neck squamous cell carcinoma. Oncol Lett 2018;16:4179-84. [Crossref] [PubMed]
- Wu Q, Zhao Y, Wang P. miR-204 inhibits angiogenesis and promotes sensitivity to cetuximab in head and neck squamous cell carcinoma cells by blocking JAK2-STAT3 signaling. Biomed Pharmacother 2018;99:278-85. [Crossref] [PubMed]
- Zhuang Z, Yu P, Xie N, et al. MicroRNA-204-5p is a tumor suppressor and potential therapeutic target in head and neck squamous cell carcinoma. Theranostics 2020;10:1433-53. [Crossref] [PubMed]
- Shi W, Yang J, Li S, et al. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget 2015;6:40172-85. [Crossref] [PubMed]
- Luo J, Wu J, Li Z, et al. miR-375 Suppresses IGF1R Expression and Contributes to Inhibition of Cell Progression in Laryngeal Squamous Cell Carcinoma. Biomed Res Int 2014;2014:374598. [Crossref] [PubMed]
- Wu Q, Zhao Y, Sun Y, et al. miR-375 inhibits IFN-γ-induced programmed death 1 ligand 1 surface expression in head and neck squamous cell carcinoma cells by blocking JAK2/STAT1 signaling. Oncol Rep 2018;39:1461-8. [PubMed]
- Ma HF, Lv GX, Zhang DH. miR-381 Mediates the Development of Head and Neck Squamous Cell Carcinoma via Targeting STC2. Onco Targets Ther 2020;13:4485-93. [Crossref] [PubMed]
- Li Y, Tao C, Dai L, et al. MicroRNA-625 Inhibits Cell Invasion and Epithelial-Mesenchymal Transition by Targeting SOX4 in Laryngeal Squamous Cell Carcinoma. Biosci Rep 2019;39:BSR20181882. [Crossref] [PubMed]
- Blandino G, Di Agostino S. New therapeutic strategies to treat human cancers expressing mutant p53 proteins. J Exp Clin Cancer Res 2018;37:30. [Crossref] [PubMed]
- Ganci F, Sacconi A, Bossel Ben-Moshe N, et al. Expression of TP53 mutation associated microRNAs predicts clinical outcome in head and neck squamous cell carcinoma patients. Ann Oncol 2013;24:3082-8. [Crossref] [PubMed]
- Nagai H, Hasegawa S, Uchida F, et al. MicroRNA-205-5p suppresses the invasiveness of oral squamous cell carcinoma by inhibiting TIMP 2 expression. Int J Oncol 2018;52:841-50. [Crossref] [PubMed]
- Childs G, Fazzari M, Kung G, et al. Low-level Expression of microRNAs let-7d and miR-205 Are Prognostic Markers of Head and Neck Squamous Cell Carcinoma. Am J Pathol 2009;174:736-45. [Crossref] [PubMed]
- Chen D, Cabay RJ, Jin Y, et al. MicroRNA Deregulations in Head and Neck Squamous Cell Carcinomas. J Oral Maxillofac Res 2013;4:e2. [Crossref] [PubMed]
- Di Agostino S, Valenti F, Sacconi A, et al. Long Non-coding MIR205HG Depletes Hsa-miR-590-3p Leading to Unrestrained Proliferation in Head and Neck Squamous Cell Carcinoma. Theranostics 2018;8:1850-68. [Crossref] [PubMed]
- Fadhil RS, Wei MQ, Nikolarakos D, et al. Salivary microRNA miR-let-7a-5p and miR-3928 could be used as potential diagnostic bio-markers for head and neck squamous cell carcinoma. PLoS One 2020;15:e0221779. [Crossref] [PubMed]
- Kent OA, McCall MN, Cornish TC, et al. SURVEY AND SUMMARY Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res 2014;42:7528-38. [Crossref] [PubMed]
- Koshizuka K, Hanazawa T, Fukumoto I, et al. The microRNA signatures: aberrantly expressed microRNAs in head and neck squamous cell carcinoma. J Hum Genet 2017;62:3-13. [Crossref] [PubMed]
- Mataki H, Seki N, Mizuno K, et al. Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p) coordinately targeted MTDH in lung squamous cell carcinoma. Oncotarget 2016;7:72084-98. [Crossref] [PubMed]
- Matsushita R, Yoshino H, Enokida H, et al. Regulation of UHRF1 by dual-strand tumor-suppressor microRNA-145 (miR-145-5p and miR-145-3p): inhibition of bladder cancer cell aggressiveness. Oncotarget 2016;7:28460-87. [Crossref] [PubMed]
- Kilic A, Barlak N, Sanli F, et al. Mode of Action of Carboplatin Via Activating p53/miR-145 Axis in Head and Neck Cancers. Laryngoscope 2020;130:2818-24. [Crossref] [PubMed]
- Harris T, Jimenez L, Kawachi N, et al. Low-Level Expression of miR-375 Correlates with Poor Outcome and Metastasis While Altering the Invasive Properties of Head and Neck Squamous Cell Carcinomas. Am J Pathol 2012;180:917-28. [Crossref] [PubMed]
- Wang P, Xu LL, Ren SS, et al. The microRNA-375 as a potentially promising biomarker to predict the prognosis of patients with head and neck or esophageal squamous cell carcinoma: a meta-analysis. Eur Arch Otorhinolaryngol 2019;276:957-68. [Crossref] [PubMed]
- Siow MY, Karen Ng LP, Vincent Chong VK, et al. Dysregulation of miR-31 and miR-375 expression is associated with clinical outcomes in oral carcinoma. Oral Dis 2014;20:345-351. [Crossref] [PubMed]
- Hu A, Huang JJ, Xu WH, et al. MiR-21/miR-375 ratio is an independent prognostic factor in patients with laryngeal squamous cell carcinoma. Am J Cancer Res 2015;5:1775-85. [PubMed]
- Khalil DN, Smith EL, Brentjens RJ. The Future of Cancer Treatment: Immunomodulation, CARs and Combination Immunotherapy. Nat Rev Clin Oncol 2016;13:273-90. [Crossref] [PubMed]
- Yang CX, Sedhom W, Song J, et al. The Role of MicroRNAs in Recurrence and Metastasis of Head and Neck Squamous Cell Carcinoma. Cancers 2019;11:395. [Crossref] [PubMed]
- Kalluri R, Weinberg A. The basics of epithelial-mesenchymal transition. J Clin Invest 2009;119:1420-8. [Crossref] [PubMed]
- Abba ML, Patil N, Leupold JH, et al. MicroRNA Regulation of Epithelial to Mesenchymal Transition. J Clin Med 2016;5:8. [Crossref] [PubMed]
- Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and Metastasis. Cancer Metastasis Rev 2012;31:653-62. [Crossref] [PubMed]
- Park SM, Gaur AB, Lengyel E, et al. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 2008;22:894-907. [Crossref] [PubMed]
- Tamagawa S, Beder LB, Hotomi M, et al. Role of miR-200c/miR-141 in the Regulation of Epithelial-Mesenchymal Transition and Migration in Head and Neck Squamous Cell Carcinoma. Int J Mol Med 2014;33:879-86. [Crossref] [PubMed]
- Díaz-Martín J, Díaz-López A, Moreno-Bueno G, et al. A Core microRNA Signature Associated With Inducers of the Epithelial-To-Mesenchymal Transition. J Pathol 2014;232:319-29. [Crossref] [PubMed]
- Koshizuka K, Hanazawa T, Arai T, et al. Involvement of aberrantly expressed microRNAs in the pathogenesis of head and neck squamous cell carcinoma. Cancer Metastasis Rev 2017;36:525-45. [Crossref] [PubMed]
- Lo WL, Yu CC, Chiou GY, et al. MicroRNA-200c attenuates tumour growth and metastasis of presumptive head and neck squamous cell carcinoma stem cells. J Pathol 2011;223:482-95. [Crossref] [PubMed]
- Zeng D, Xu H, Ji N, et al. In situ measurement of miR-138 expression in oral squamous cell carcinoma tissue supports the role of this microRNA as a tumor suppressor. J Oral Pathol Med 2019;48:911-8. [Crossref] [PubMed]
- Liu X, Jiang L, Wang A, et al. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett 2009;286:217-22. [Crossref] [PubMed]
- Ji M, Wang W, Yan W, et al. Dysregulation of AKT1, a miR-138 target gene, is involved in the migration and invasion of tongue squamous cell carcinoma. J Oral Pathol Med 2017;46:731-7. [Crossref] [PubMed]
- Liu X, Wang C, Chen Z, et al. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J 2011;440:23-31. [Crossref] [PubMed]
- Jiang L, Liu X, Kolokythas A. Down-regulation of the Rho GTPase signaling pathway is involved in the microRNA-138 mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer 2010;127:505-12. [Crossref] [PubMed]
- Bao L, Miao W, Yu Y. Reduced serum miR-138 is associated with poor prognosis of head and neck squamous cell carcinoma. Int J Clin Exp Pathol 2017;10:10276-81. [PubMed]
- Kim H, Yang JM, Ahn SH, et al. Potential Oncogenic Role and Prognostic Implication of MicroRNA-155-5p in Oral Squamous Cell Carcinoma. Anticancer Research 2018;38:5193-200. [Crossref] [PubMed]
- Faraoni I, Antonetti FR, Cardone J, et al. miR-155 gene: A typical multifunctional microRNA. Biochimica et Biophysica Acta 2009;1792:497-505. [Crossref] [PubMed]
- Baba O, Hasegawa S, Nagai H, et al. MicroRNA-155-5p is associated with oral squamous cell carcinoma metastasis and poor prognosis. J Oral Pathol Med 2016;45:248-55. [Crossref] [PubMed]
- Lubov J, Maschietto M, Ibrahim I, et al. Meta-analysis of microRNAs expression in head and neck cancer: uncovering association with outcome and mechanisms. Oncotarget 2017;8:55511-24. [Crossref] [PubMed]
- Arunkumar G, Deva Magendhra Rao AK, Manikandan M, et al. Dysregulation of miR 200 family microRNAs and epithelial mesenchymal transition markers in oral squamous cell carcinoma. Oncol Lett 2018;15:649-57. [PubMed]
- Ganci F, Sacconi A, Manciocco V, et al. MicroRNA Expression as Predictor of Local Recurrence Risk in Oral Squamous Cell Carcinoma. Head Neck 2016;38:E189-97. [Crossref] [PubMed]
- Ganci F, Sacconi A, Manciocco V, et al. Altered Peritumoral microRNA Expression Predicts Head and Neck Cancer Patients With a High Risk of Recurrence. Mod Pathol 2017;30:1387-401. [Crossref] [PubMed]
- Cohen EEW, Soulières D, Le Tourneau C, et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): a randomised, open-label, phase 3 study. Lancet 2019;393:156-67. [Crossref] [PubMed]
- Burtness B, Harrington KJ, Greil R, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet 2019;394:1915-28. [Crossref] [PubMed]
- Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer 2018;18:269-282. [Crossref] [PubMed]
- Perri F, Pacelli R, Della Vittoria Scarpati G, et al. Radioresistance in head and neck squamous cell carcinoma: Biological bases and therapeutic implications. Head Neck 2015;37:763-70. [Crossref] [PubMed]
- Cellini F, Morganti AG, Genovesi D, et al. Role of microRNA in response to ionizing radiations: evidences and potential impact on clinical practice for radiotherapy. Molecules 2014;19:5379-401. [Crossref] [PubMed]
- Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev 2013;23:12-9. [Crossref] [PubMed]
- Ganci F, Sacconi A, Manciocco V, et al. Radioresistance in head and neck squamous cell carcinoma—possible molecular markers for local recurrence and new putative therapeutic strategies. In: Contemporary Issues in Head and Neck Cancer Management. London, UK: InTech, 2015.
- Ahmad P, Sana J, Slavik M, et al. MicroRNAs Involvement in Radioresistance of Head and Neck Cancer. Dis Markers 2017;2017:8245345. [Crossref] [PubMed]
- Koshizuka K, Hanazawa T, Fukumoto I, et al. Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum Genet 2017;62:113-21. [Crossref] [PubMed]
- Yang CH, Wang Y, Sims M, et al. MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer Lett 2019;465:59-67. [Crossref] [PubMed]
- Mataki H, Enokida H, Chiyomaru T, et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin1C. J Hum Genet 2015;60:53-61. [Crossref] [PubMed]
- Migliore C, Martin V, Leoni VP, et al. MiR-1 downregulation cooperates with MACC1 in promoting MET overexpression in human colon cancer. Clin Cancer Res 2012;18:737-47. [Crossref] [PubMed]
- Nohata N, Hanazawa T, Enokida H, et al. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget 2012;3:9-21. [Crossref] [PubMed]
- Zhang L, Liu X, Jin H, et al. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett 2013;332:94-101. [Crossref] [PubMed]
- Vermorken JB, Mesia R, Rivera F, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med 2008;359:1116-27. [Crossref] [PubMed]
- Siano M, Espeli V, Mach N, et al. Gene signatures and expression of miRNAs associated with efficacy of panitumumab in a head and neck cancer phase II trial. Oral Oncol 2018;82:144-51. [Crossref] [PubMed]
- Bossi P, Bergamini C, Siano M, et al. Functional Genomics Uncover the Biology behind the Responsiveness of Head and Neck Squamous Cell Cancer Patients to Cetuximab. Clin Cancer Res 2016;22:3961-70. [Crossref] [PubMed]
- De Cecco L, Giannoccaro M, Marchesi E, et al. Integrative miRNA-gene expression analysis enables refinement of associated biology and prediction of response to cetuximab in head and neck squamous cell cancer. Genes (Basel) 2017;8:35. [Crossref] [PubMed]
- Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) 2017;9:52. [Crossref] [PubMed]
- Wu C, Tong L, Wu C, et al. Two miRNA prognostic signatures of head and neck squamous cell carcinoma: A bioinformatic analysis based on the TCGA dataset. Cancer Med 2020;9:2631-42. [Crossref] [PubMed]
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 2007;67:7713-22. [Crossref] [PubMed]
- Tran N, McLean T, Zhang X, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun 2007;358:12-7. [Crossref] [PubMed]
- Manikandan M, Deva Magendhra Rao AK, Arunkumar G, et al. Oral squamous cell carcinoma: microRNA expression profiling and integrative analyses for elucidation of tumourigenesis mechanism. Mol Cancer 2016;15:28. [Crossref] [PubMed]
- Xie Z, Cao L, Zhang J. miR-21 modulates paclitaxel sensitivity and hypoxia-inducible factor-1α expression in human ovarian cancer cells. Oncol Lett 2013;6:795-800. [Crossref] [PubMed]
- Chan JK, Blansit K, Kiet T, et al. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol Oncol 2014;132:739-44. [Crossref] [PubMed]
- Feng YH, Tsao CJ. Emerging role of microRNA-21 in cancer. Biomed Rep 2016;5:395-402. [Crossref] [PubMed]
- Au Yeung CL, Co NN, Tsuruga T, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun 2016;7:11150. [Crossref] [PubMed]
- Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157-60. [Crossref] [PubMed]
- Zhou G, Liu Z, Myers JN. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J Cell Biochem 2016;117:2682-92. [Crossref] [PubMed]
- Fu X, Han Y, Wu Y, et al. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest 2011;41:1245-53. [Crossref] [PubMed]
- Liu B, Su F, Li Y, et al. Changes of serum miR34a expression during neoadjuvant chemotherapy predict the treatment response and prognosis in stage II/III breast cancer. Biomed Pharmacother 2017;88:911-7. [Crossref] [PubMed]
- Zhang L, Liao Y, Tang L. MicroRNA-34 family: a potential tumor suppressor and therapeutic candidate in cancer. J Exp Clin Cancer Res 2019;38:53. [Crossref] [PubMed]
- Soltani-Sedeh H, Irani S, Mirfakhraie R, Soleimani M. Potential using of microRNA-34A in combination with paclitaxel in colorectal cancer cells. J Cancer Res Ther 2019;15:32-7. [PubMed]
- Naghizadeh S, Mohammadi A, Duijf PHG, et al. The role of miR-34 in cancer drug resistance. J Cell Physiol 2020;235:6424-40. [Crossref] [PubMed]
- Wiggins JF, Ruffino L, Kelnar K, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res 2010;70:5923-30. [Crossref] [PubMed]
- Liu C, Kelnar K, Liu B, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 2011;17:211-5. [Crossref] [PubMed]
- Peng D, Wang H, Li L, et al. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia 2018;32:1180-8. [Crossref] [PubMed]
- Kumar B, Yadav A, Lang J. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One 2012;7:e37601. [Crossref] [PubMed]
- Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet 2012;3:120. [Crossref] [PubMed]
- Beg MS, Brenner AJ, Sachdev J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 2017;35:180-8. [Crossref] [PubMed]
- Hong DS, Kang YK, Borad M, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer 2020;122:1630-7. [Crossref] [PubMed]
- Gorenc M, Kozjek NR, Strojan P. Malnutrition and cachexia in patients with head and neck cancer treated with (chemo)radiotherapy. Rep. Pract. Oncol. Radiother 2015;20:249-58. [Crossref] [PubMed]
- Boguszewicz Ł, Bieleń A, Mrochem-Kwarciak J, et al. NMR-based metabolomics in real-time monitoring of treatment induced toxicity and cachexia in head and neck cancer: a method for early detection of high risk patients. Metabolomics 2019;15:110. [Crossref] [PubMed]
- Fearon K, Strasser F, Anker SD, et al. Definition and classification of cancer cachexia: an international consensus. Lancet Oncol 2011;12:489-95. [Crossref] [PubMed]
- Baracos VE, Martin L, Korc M, et al. Cancer-associated cachexia. Nat Rev Dis Primers 2018;4:17105. [Crossref] [PubMed]
- Argilés JM, Busquets S, Stemmler B, et al. Cancer cachexia: understanding the molecular basis. Nat Rev Cancer 2014;14:754-62. [Crossref] [PubMed]
- Tseng YC, Kulp SK, Lai IL, et al. Preclinical Investigation of the Novel Histone Deacetylase Inhibitor AR-42 in the Treatment of Cancer-Induced Cachexia. J Natl Cancer Inst 2015;107:djv274. [Crossref] [PubMed]
- Freire PP, Fernandez GJ, Cury SS, et al. The Pathway to Cancer Cachexia: MicroRNA-Regulated Networks in Muscle Wasting Based on Integrative Meta-Analysis. Int J Mol Sci 2019;20:1962. [Crossref] [PubMed]
- Donzelli S, Farneti A, Marucci L, et al. Non-coding RNAs as Putative Biomarkers of Cancer-Associated Cachexia. Front Cell Dev Biol 2020;8:257. [Crossref] [PubMed]
- Santos JMO, Peixoto da Silva S, Gil da Costa RM, et al. The Emerging Role of MicroRNAs and Other Non-Coding RNAs in Cancer Cachexia. Cancers (Basel) 2020;12:1004. [Crossref] [PubMed]
- Powrózek T, Mlak R, Brzozowska A, et al. miRNA-130a Significantly Improves Accuracy of SGA Nutritional Assessment Tool in Prediction of Malnutrition and Cachexia in Radiotherapy-Treated Head and Neck Cancer Patients. Cancers (Basel) 2018;10:294. [Crossref] [PubMed]
- Zhang J, Wu H, Li P, Zhao Y, et al. NF-κB-modulated miR-130a targets TNF-α in cervical cancer cells. J Transl Med 2014;12:155. [Crossref] [PubMed]
- He WA, Calore F, Londhe P, et al. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci U S A 2014;111:4525-9. [Crossref] [PubMed]
- Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR-21 in Cancer. Drug Dev Res 2015;76:270-7. [Crossref] [PubMed]
- Valadi H, Ekström K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9. [Crossref] [PubMed]
- Hammerman PS, Hayes DN, Grandis JR. Therapeutic insights from genomic studies of head and neck squamous cell carcinomas. Cancer Discov 2015;5:239-44. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Li J, Han X, Yu X, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res 2018;37:213. [Crossref] [PubMed]
- Ozawa PMM, Jucoski TS, Vieira E, et al. Liquid biopsy for breast cancer using extracellular vesicles and cell-free microRNAs as biomarkers. Transl Res 2020;223:40-60. [Crossref] [PubMed]
- Dioguardi M, Caloro GA, Laino L, et al. Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis. Cancers (Basel) 2020;12:936. [Crossref] [PubMed]
- Li L, Li C, Wang S, et al. Exosomes Derived from Hypoxic Oral Squamous Cell Carcinoma Cells Deliver miR-21 to Normoxic Cells to Elicit a Prometastatic Phenotype. Cancer Res 2016;76:1770-80. [Crossref] [PubMed]
- Ishinaga H, He F, Hou B, et al. A longitudinal study on circulating miR-21 as a therapeutic effect marker in head and neck squamous cell carcinoma. Carcinogenesis 2019; [Crossref] [PubMed]
- Hsu CM, Lin PM, Wang YM, et al. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol 2012;33:1933-42. [Crossref] [PubMed]
- Hou B, Ishinaga H, Midorikawa K, et al. Circulating microRNAs as novel prognosis biomarkers for head and neck squamous cell carcinoma. Cancer Biol Ther 2015;16:1042-6. [Crossref] [PubMed]
- Gao S, Xu Q, Zhou Y, et al. Serum levels of microRNA-21 and microRNA-10a can predict long-term prognosis in laryngeal cancer patients: a multicenter study. Transl Cancer Res 2020;9:3680-90. [Crossref]
- Lu J, Xu X, Liu X, et al. Predictive value of miR-9 as a potential biomarker for nasopharyngeal carcinoma metastasis. Br J Cancer 2014;110:392-8. [Crossref] [PubMed]
- Mirghani H, Amen F, Tao Y, et al. Increased radiosensitivity of HPV-positive head and neck cancers: Molecular basis and therapeutic perspectives. Cancer Treat Rev 2015;41:844-52. [Crossref] [PubMed]
- Lajer CB, Garnæs E, Friis-Hansen L, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer 2017;117:e2. [Crossref] [PubMed]
- Gurin D, Slavik M, Hermanova M, et al. The Tumor Immune Microenvironment and Its Implications for Clinical Outcome in Patients with Oropharyngeal Squamous Cell Carcinoma. J Oral Pathol Med 2020;49:886-96. [Crossref] [PubMed]
- Tong F, Mao X, Zhang S, et al. HPV + HNSCC-derived exosomal miR-9 induces macrophage M1 polarization and increases tumor radiosensitivity. Cancer Lett 2020;478:34-44. [Crossref] [PubMed]
- Summerer I, Unger K, Braselmann H, et al. Circulating microRNAs as prognostic therapy biomarkers in head and neck cancer patients. Br J Cancer 2015;113:76-82. [Crossref] [PubMed]
- Greither T, Vorwerk F, Kappler M, et al. Salivary miR-93 and miR-200a as post-radiotherapy biomarkers in head and neck squamous cell carcinoma. Oncol Rep 2017;38:1268-75. [Crossref] [PubMed]
- Schneider A, Victoria B, Lopez YN, et al. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci Rep 2018;8:675. [Crossref] [PubMed]
- Citron F, Armenia J, Franchin G, et al. An Integrated Approach Identifies Mediators of Local Recurrence in Head and Neck Squamous Carcinoma. Clin Cancer Res 2017;23:3769-80. [Crossref] [PubMed]
- Cramer JD, Burtness B, Ferris RL. Immunotherapy for head and neck cancer: Recent advances and future directions. Oral Oncol 2019;99:104460. [Crossref] [PubMed]
- von der Grün J, Rödel F, Brandts C, et al. Targeted Therapies and Immune-Checkpoint Inhibition in Head and Neck Squamous Cell Carcinoma: Where Do We Stand Today and Where to Go? Cancers (Basel) 2019;11:472. [Crossref] [PubMed]
- Pesce S, Greppi M, Ferretti E, et al. miRNAs in NK Cell-Based Immune Responses and Cancer Immunotherapy. Front Cell Dev Biol 2020;8:119. [Crossref] [PubMed]
- Omar HA, El-Serafi AT, Hersi F, et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J 2019;286:3540-57. [Crossref] [PubMed]
- Yu D, Liu X, Han G, et al. The let-7 family of microRNAs suppresses immune evasion in head and neck squamous cell carcinoma by promoting PD-L1 degradation. Cell Commun Signal 2019;17:173. [Crossref] [PubMed]
- Neoadjuvant Nivolumab for Oral Cancer Combined with FDG and Anti-PD-L1 PET/CT Imaging for Response Prediction-ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03843515
- FRAIL-IMMUNE (GORTEC 2018-03)-Combination of Durvalumab With Carboplatin/Paclitaxel—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT03723967


