
Chromodomain-helicase-DNA-binding protein 1-like (CHD1L) silencing inhibits gastric cancer cell proliferation, invasion, and migration
Introduction
Gastric cancer (GC) is second most common malignancy worldwide, with more than 1,000,000 new cases and an estimated 783,000 deaths in 2018 (equaling 1 out of every 12 deaths globally) (1). There is a high incidence of GC in China, with 436,000 new cases and 323,000 deaths reported in 2017 (2). GC arises via multi-step, multi-factor processes, including deficiencies of tumor suppressor genes or the activation of oncogenes and associated transfer genes (3). For proper diagnosis, staging, and prognosis of GC, it is essential to understand cellular proliferation and the activation and expression of key genes in GC.
In 1991, the chromodomain-helicase-DNA-binding protein 1-like (CHD1L) gene was first isolated by Ma et al. (4) from the 1q21 region of the long arm of chromosome 1 from 60 patients with primary hepatocellular carcinoma. CHD1L contains 23 exons, an open reading frame encoding 897 amino acids, and a conserved region (5). CHD1L belongs to the sucrose non-fermentable (SNF)-2 family and contains a 400-amino acid domain in its functional structure that is similar to other members of the SNF-2 family, namely SWItch and CHD1 (6). However, CHD1L shows limited conserved helicase function and does not recognize the terminal chromosome structure of methylated histones (7). Similar to the important roles played by SNF-2 family members, including regulation of the binding of DNA and proteins, regulation of transcription, and recombination of chromosomes, CHD1L plays pivotal roles in chromatin remodeling and regulation of protein-DNA binding. The main mechanisms of action of CHD1L include induction of chromosomal instability and mitosis, inhibition of apoptosis, regulation of the cell cycle, and participation in DNA damage and repair processes (8). Additionally, CHD1L binds to nerve growth factor IB (Nur77), inhibiting its migration from the nucleus to the mitochondria and blocking the release of cytochrome c into the cytoplasm, which in turn inhibits the activity of caspase 3 and 9, ultimately inhibiting apoptosis (9). ARHGEF9 belongs to the Rho-like GTPase family (Ras homolog) and functions as a molecular switch in the conversion of active GTP to inactive GDP (10). ARHGEF9-encoded actin plays a key role in the cytoskeleton and cell signaling pathways (11). CHD1L and ARHGEF9 encode effector molecules with inhibitory effects on the occurrence and development of tumors that play a key role in the development of GC (12,13).
CHD1L overexpression leads to over-relaxation of chromatin, causing DNA base mismatch and development of cancer (14). CHD1L is highly expressed in colorectal cancer, GC, ovarian cancer, bladder cancer, breast cancer, nasopharyngeal cancer, lung adenocarcinoma, and glioma (15). CHD1L overexpression significantly promotes the proliferation, invasion, and migration of colon cancer cells, whereas its silencing reduces the invasion and metastasis of prostate cancer cells (5,16). However, it remains unknown if CHD1L promotes the proliferation, apoptosis, invasion, and metastasis of GC cells. In GC, CHD1L is a relatively novel gene, and its role in the occurrence and development of GC remains under investigation. As such, it was hypothesized that inhibition or silencing of CHD1L may be a novel approach for the treatment of GC and may improve the effects of radio-chemotherapy and gene therapy and the prognosis of GC patients. In this study, a stable lentiviral vector carrying CHD1L small hairpin RNA (shRNA) was established and its effect on the biological functions of GC cells was evaluated by MTT and Transwell assays. CHD1L was found to play a crucial role in several tumor cell activities, including migration, proliferation, and apoptosis. This study focused on a systematic multi-aspect analysis of CHD1L in terms of the occurrence and development of GC. Furthermore, the molecular mechanism of GC was evaluated from multiple angles, and CHD1L was found to be regulate GC progression. These results may provide a basis for the development of novel anticancer drugs targeting CHD1L. We present the following article in accordance with the MDAR Reporting Checklist (available at http://dx.doi.org/10.21037/tcr-19-2700).
Methods
Reagents and materials
The pLVshRNA-Puro lentivirus vector was provided by Clontech Laboratories, Inc. The RNA extraction reagent TRIzol® and the Annexin V-FITC/PI double-staining cell apoptosis detection kit were provided by Invitrogen (Thermo Fisher Scientific, Inc.). The MTT and BCA kits were provided by Sigma-Aldrich; Merck KGaA. DMEM, trypsin, FBS, and cell culture reagents were provided by Gibco (Thermo Fisher Scientific, Inc.). Rabbit anti-human FITC-conjugated IgG (cat. no. sc2778), mouse anti-human CHD1L (cat. no. sc81065), and the ECL chemiluminescence kit were provided by Santa Cruz Biotechnology, Inc.
shRNA design and preparation
The CHD1L mRNA sequence was downloaded from the GenBank database (http://www.exiqon.com/), and corresponding shRNAs were designed. Primers used to prepare shRNAs are listed in Table 1. shRNA sequences were confirmed to be free from other homologous sequences using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). shRNAs were cloned into the pLVshRNA-Puro vector by digestion with EcoRI and NotI. Successful clones were identified by sequencing. The knockdown pLVshRNA-Puro-CHD1L (CHD1L-shRNA) and the negative control (NC) pLVshRNA-Puro-NC (shRNA-NC) were packaged according to the viral packaging protocol and provided by Clontech Laboratories, Inc. Viral supernatants were collected after 48 and 72 h, followed by centrifugation (50,000 ×g; 4 °C; 4 h) and filtration. The filtrate was concentrated at 4 °C at 50,000 ×g for 4 h to 108/mL and stored at −80 °C.
Table 1
| Name | Sequence (5'-3') |
|---|---|
| CHD1L-shRNA-1-F | CCUGCUGGAUAAGCUACUATT |
| CHD1L-shRNA-1-R | UAGUAGCUUAUCCAGCAGGTT |
| CHD1L-shRNA-2-F | GGAGACUCAUAGAGGAGAATT |
| CHD1L -shRNA-2-R | UUCUCCUCUAUGAGUCUCCTT |
| CHD1L-shRNA-3-F | GGCAUCCCAACUUACAUAUTT |
| CHD1L-shRNA-3-R | AUAUGUAAGUUGGGAUGCCTT |
| NC-F | UUCUUGAACGUGUCACGUTT |
| NC-R | AUGUGACACGUUCGGAGAATT |
CHD1L, chromodomain-helicase-DNA-binding protein 1-like; NC, negative control; F, forward; R, reverse.
Infection of BGC-823 cells with lentiviral shRNAs and the establishment of stable clones
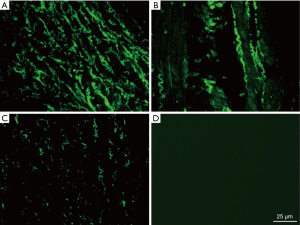
The GC cell line BGC-823 (Type Culture Collection of the Chinese Academy of Sciences) was cultured in DMEM containing 10% FBS, 100 µg/mL streptomycin, and 100 U/mL penicillin at 37 °C with 5% CO2. The medium was replaced every 2–3 d. BGC-823 cells in the logarithmic growth phase were seeded in 6-well plates (2×105/well) and infected with 0.5 mL of viral supernatant [multiplicity of infection (MOI), 150] containing CHD1L-shRNA-1 to -3 or shRNA-NC for 48 h. Cells were then diluted 1:20, and puromycin (0.25 µg/mL) was added after 24 h. Puromycin-resistant cells were collected after 48 h and assessed for GFP fluorescence using a fluorescence microscope (magnification, ×100) in the 3rd, 15th and 25th generation.
Screening of CHD1L silencing
Lentivirus-infected BGC-823 cells were harvested and rinsed three times, and total protein was extracted using RIPA-buffer. Protein concentration was measured using the BCA protein concentration detection kit. Proteins (60 µg) were isolated by 10% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked in 5% skim milk powder-TBST for 2 h at room temperature and incubated with primary anti-CHD1L antibody (1:1,000; cat. no. sc81065) and monoclonal β-actin antibody (1:1,1000; cat. no. sc58673; Santa Cruz Biotechnology, Inc.) at 4 °C overnight. Membranes were rinsed with TBST three times and incubated with secondary antibody [1:2,000; HRP-conjugated-goat anti-rabbit polyclonal IgG; cat. no. D110058, Sangon Biotech (Shanghai) Co., Ltd., China] at 37 °C for 2 h. An ECL chemiluminescence kit was used to visualize protein bands. Images were analyzed with Image-Pro Plus 8.0 software (Media Cybernetics, Inc.), and the ratio of CHD1L to β-actin was determined. The CHD1L-shRNA with the highest knockdown efficiency was used in subsequent experiments.
Western blot
Expression of p53 (cat. no. sc377567), p21 (cat. no. sc166630), Nur77 (cat. no. sc365113), and ARHGEF9 (cat. no. sc136393) was detected by western blotting in BGC-823 cells infected with CHD1L-shRNA-1 or shRNA-NC and in untreated cells. Proteins were isolated as described above. Membranes were incubated overnight at 4 °C with primary antibodies (1:1,000) and at 37 °C for 2 h with the anti-β-actin antibody (1:2,000; cat. no. sc58673). Membranes were then incubated with secondary antibody (HRP-conjugated goat anti-mouse IgG; cat. no. A21010; 1:5,000, Abbkine, Wuhan, China) at 37 °C for 2 h. Bands were visualized and analyzed as described above. Experiments were repeated three times.
MTT assay
BGC-823 cells infected with CHD1L-shRNA-1 or shRNA-NC or untreated cells were seeded in 96-well plates at 1×103 cells/well. Cells were incubated at 37 °C with 5% CO2 for 24 h, then 10 µL MTT solution (5 mg/mL) was added, and cells were incubated for an additional 4 h. After removing the supernatant, DMSO (150 µL/well) was added, plates were gently mixed for 10 min at 37 °C, and the absorbance at 490 nm was measured. Cell growth curves were prepared at 0, 12, 24, 36, 48, 60, and 72 h. Experiments were repeated three times.
Flow cytometry
After 48 h of trypsin digestion, BGC-823 cells infected with CHD1L-shRNA-1 or shRNA-NC, or untreated cells were rinsed with PBS, and the supernatant was removed by centrifugation (4 °C; 50,000 ×g; 5 min). After repeating this twice, cells were cultured for 24 h, and single-cell suspensions were prepared and collected for staining following the instructions of the cell cycle detection and apoptosis detection kit for flow cytometry. Cells (5×105) were collected, and Annexin V-FITC/PI (Invitrogen; Thermo Fisher Scientific, Inc.) was added, gently mixed, and incubated at room temperature for 15 min. Cell suspensions were analyzed using a flow cytometer within 1 h.
Transwell invasion assay
Single-cell suspensions (100 µL) of BGC-823 cells infected with CHD1L-shRNA-1 or shRNA-NC or untreated cells (5×105 cells/mL in serum-free medium) and 500 µL fibronectin (PHE0023; 80 ng/mL; Thermo Fisher Scientific, Inc.) were added to the upper and lower Matrigel-coated Transwell chambers, respectively, and cultured at 37 °C with 5% CO2 for 24 h. After removing the Matrigel and non-invasive cells from the membrane, cells were fixed in precooled methanol for 30 min at 4 °C, followed by 5 min of Giemsa staining (1 mg/mL) at room temperature, ethanol dehydration (80%, 95%, and 97%), and gradient elution with a 20 min incubation at room temperature for each step. The number of invasive cells in five randomly selected high-power microscopic fields (magnification, ×200) was counted under an inverted microscope. The inhibitory rate of CHD1L silencing in GC cells was calculated as follows: inhibitory rate (%) = (control − sample)/control ×100.
Wound healing assay
BGC-823 cells infected with CHD1L-shRNA-1 or shRNA-NC or untreated cells (5×105) were seeded in 6-well plates and incubated for 24 h to reach 97% confluence. Vertical lines were drawn with a pipette tip (200 µL), and loose cells were washed away using PBS. The plates were incubated with serum-free medium at 37 °C with 5% CO2 and photographed using an inverted microscope (magnification, ×100) at 0 and 24 h post wounding. The intercellular distance was calculated for further comparison using ImageJ (1.48; National Institutes of Health). For data processing, the distance at 24 h post wounding was compared with the starting distance at 0 h post wounding.
Statistical analysis
SPSS19.0 (IBM Corp.) was used for statistical analysis. Data are expressed as the mean ± standard deviation, and experiments were repeated three times. Normally distributed and homogeneous data were compared using one-way ANOVA with least significant difference test. Other data were compared using the Wilcoxon rank sum test. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of CHD1L knockdown cells
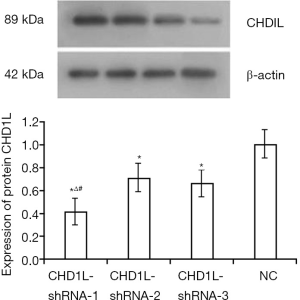
Upon infection of GC cells with CHD1L-shRNA-1 to -3 and shRNA-NC, cellular morphology did not change significantly (data not shown). At 48 h after infection, expression of fluorescent protein was ~50% with no significant decrease in expression in the 3rd, 15th, and 25th generation of cells in the CHD1L-shRNA-1 group. This indicated that the lentivirus was safely and stably integrated into the genome of target cells and achieved long-term expression of the CHD1L-shRNAs (Figure 1). As shown in Figure 2, compared with the shRNA-NC group, CHD1L expression in the CHD1L-shRNA-1, -2, and -3 groups was significantly decreased by 58.60%, 28.99%, and 33.49%, respectively. Given that CHD1L-shRNA-1 allowed the strongest silencing of CHD1L, this group was selected for subsequent analyses.
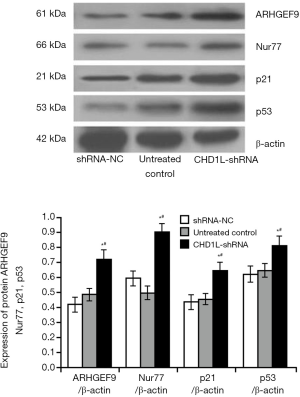
CHD1L silencing increases the expression of growth and apoptosis associated proteins
As shown in Figure 3, the expression of p53, p21, Nur77, and ARHGEF9 expression, associated with growth and apoptosis inhibition in BGC-823 cells (17,18), was significantly increased by 31.24%, 48.37%, 52.36%, and 49.78%, respectively, in CHD1L-shRNA-1-treated cells compared with shRNA-NC-treated cells (P=0.041, 0.030, 0.026, and 0.019, respectively). This indicated that CHD1L silencing promotes the expression of factors that inhibit tumor occurrence and development.
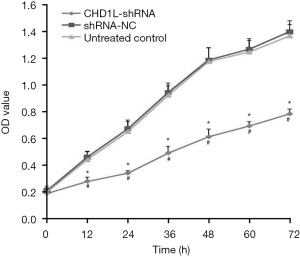
CHD1L silencing decreases proliferation
Results of the MTT assays revealed that CHD1L-shRNA-1 was effective as early as 12 h after transfection, and the effect persisted thereafter. Compared with shRNA-NC treated cells, the proliferation of BGC-823 cells infected with CHD1L-shRNA-1 was significantly inhibited at 12, 24, 36, 48, 60, and 72 h (P<0.05; Figure 4). The inhibitory effect was time-dependent, and the inhibitory rate rose over time. There was no statistically significant difference between the shRNA-NC-treated and untreated control groups (P>0.05), indicating that silencing of CHD1L has a significant inhibitory effect on the proliferation of BGC-823 cells.
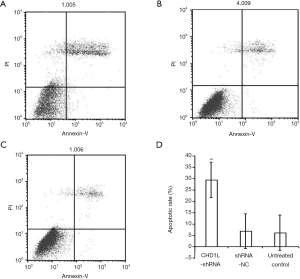
CHD1L silencing induces apoptosis and cell cycle distribution
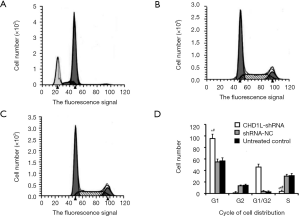
In flow cytometry analysis, cells in the lower right quadrant represent early apoptotic cells while cells in the upper right quadrant represent those in necrotic or advanced apoptotic states. Here, flow cytometry revealed that the number of apoptotic cells in the CHD1L-shRNA-1-treated cells was significantly increased 48 h after infection when compared with the shRNA-NC-treated and untreated control groups (P<0.05; Figure 5). There was no significant difference between the shRNA-NC-treated and untreated control groups (P>0.05), indicating that CHD1L silencing induced apoptosis in BGC-823 cells. Cell cycle analysis demonstrated the number of cells in the G1 phase was significantly increased in CHD1L-shRNA-1-treated cells compared to shRNA-NC-treated cells 48 h after infection (P=0.016; Figure 6), and the number of cells in the S phase was significantly decreased in CHD1L-shRNA-1-treated cells (P=0.028). These results indicated that CHD1L silencing decreases the proliferation of BGC-823 cells and induces cell cycle arrest in the G1 phase.
CHD1L silencing inhibits cell invasion
Results of the Transwell invasion assays are shown in Table 2. At 48 h, the number of invaded cells in the CHD1L-shRNA-1 group was significantly reduced compared to that in the shRNA-NC group (P=0.042), with an inhibitory rate of 59.55%. These results indicated that CHD1L silencing significantly inhibits the invasion of BGC-823 cells, and no significant difference was observed between the shRNA-NC-treated and untreated control groups (P>0.05).
Table 2
| Group | Invaded cells | Inhibition rate (%) |
|---|---|---|
| CHD1L-shRNA-1 | 58.63±10.97 | – |
| shRNA-NC | 144.95±12.68a | 59.55 |
| Untreated control | 148.49±17.86a | 60.52 |
Inhibitory rate (%) = (control − sample)/control ×100. a, P<0.05 vs. CHD1L-shRNA-1. CHD1L, chromodomain-helicase-DNA-binding protein 1-like; NC, negative control.
CHD1L silencing inhibits migration
The wound healing assay revealed that 24 h after wounding, the wounds in the shRNA-NC and untreated control groups were almost closed; however, this was not the case in the CHD1L-shRNA-1 group (Figure 7). After 24 h, the migration distance of CHD1L-shRNA-1 treated BGC-823 cells was 0.54±0.34 µm, which was significantly less than that of the shRNA-NC treated and untreated control cells (1.34±0.26 and 1.31±0.31 µm, respectively; P<0.05; Table 3). These results indicated that CHD1L silencing may significantly decrease or inhibit the migration of GC cells.
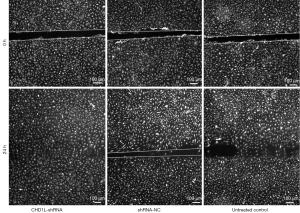
Table 3
| Group | Migration distance (μm) |
|---|---|
| CHD1L-shRNA-1 | 0.54±0.34 |
| shRNA-NC | 1.34±0.26a |
| Untreated control | 1.31±0.31a |
a, P<0.05 vs. CHD1L-shRNA-1. CHD1L, chromodomain-helicase-DNA-binding protein 1-like; NC, negative control.
Discussion
CHD1L is the only SNF-2 family gene reported to be carcinogenic (19-21). The mechanisms by which CHD1L exerts its carcinogenic effects, including tumor growth and metastasis, rely on three aspects: (I) expression of CHD1L downregulates the tumor suppressor gene p53 and its cell cycle-dependent kinase inhibitor 1A. This leads to the rapid activation of cyclin E and its dependent kinase 2, thereby inactivating phosphorylated retinoblastoma proteins in G1/S phases and accelerating the release of E2F transcription factor to ultimately induce a rapid change to the S phase and regulate apoptosis and proliferation by promoting DNA synthesis (22,23). (II) Interaction with Nur77 inhibits the migration of Nur77 from the nucleus to the mitochondria, the release of cytochrome C, the activity of caspase 3 and 9, and apoptosis (12). (III) Activation of 42 genes in the cell-division cycle induces the formation of filopodia and markedly enhances cell mobility, thus promoting epithelial-to-mesenchymal transition (EMT) and improving the metastasis, invasion, and migration of tumors (24).
CHD1L has been reported to be overexpressed in a variety of tumors and has a strong association with tumor prognosis (7,9,25-30). The mechanisms of CHD1L-involved occurrence and development of liver cancer have been investigated (31). Based on these findings, we hypothesized that the association between CHD1L and GC development may be similar. One clinical study reported that CHD1L occurrence in patients is correlated with clinical stage and distant metastasis and is an independent prognostic marker for overall survival (32). We assessed cell proliferation and found that silencing of CHD1L significantly inhibited the proliferation of BGC-823 cells, suggesting that CHD1L silencing may suppress DNA synthesis in cancer cells by promoting p53 and p21 expression. CHD1L may activate the proliferation of a large number of tumor cells, indicating that it may play a role in the occurrence and development of GC. CHD1L may be a novel precancerous marker for GC and provide novel targets for clinical gene therapies.
We further found that CHD1L silencing induces apoptosis in BGC-823 cells, suggesting that it may regulate cellular function at the transcription level. Additionally, CHD1L induces relocation of Nur77 from the nucleus to the mitochondria, thus inhibiting malignancies (33). At 48 h after CHD1L-shRNA-1 infection, the number of BGC-823 cells in the G1 phase was significantly increased, while the number of cells in the S phase was significantly decreased when compared with the shRNA-NC group, indicating that the cell cycle of BGC-823 cells was arrested in the G1 phase. Recently, CHD1L has been shown to regulate the G1/S transition and inhibit apoptosis (6,14,34,35), consistent with the results of the current study.
EMT plays an important role in tumor invasion and metastasis and is a major factor in malignant tumor-induced death (36). Therefore, any biomolecules involved in EMT that activate signal transduction pathways are likely to be key players in tumor metastasis (37). The occurrence and development of GC occur via a complex multi-step process regulated by a variety of cytokines, and EMT is a biological process that is highly conserved and prevalent in mammals (38). Under the stimulation of external physical and chemical factors, EMT is activated such that cell adhesion is weakened while migratory capacity is enhanced (39). In this study, we investigated the impact of CHD1L silencing on the migration and invasion of GC cells and found that the number of invasive cells and the migration distance in the CHD1L-shRNA group were significantly lower than those in the shRNA-NC group. ARHGEF9 is a member of the Rho family and plays a key role in regulating signaling pathways and the actin cytoskeleton (13). We hypothesized that CHD1L silencing may upregulate the expression of ARHGEF9 and Cdc42-GTP, inhibit the pseudo-like structure formation by regenerated actin, and increase cell adhesion, thus inhibiting distant metastasis, invasion, and EMT of GC cells (40).
Conclusions
CHD1L plays a role in a series of tumor cell activities, such as migration, proliferation, and apoptosis. Therefore, investigating the impact of CHD1L silencing on the biological behavior of GC cells may provide insights into the molecular mechanisms underlying the occurrence and development of GC. Future work may discuss the upstream and downstream regulatory genes of CHD1L, as well as potentially involved signaling pathways. Furthermore, it seems worthwhile to design small molecules based on the molecular structure of CHD1L to inhibit its activities, reduce the expression of CHD1L in GC cells using gene therapies, or inhibit the invasion and metastasis of GC cells to prevent recurrence of GC and develop novel CHD1L-based anticancer drugs.
Acknowledgments
Funding: The current study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-19-2700
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-19-2700
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-19-2700). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our study has not been involved in animal and human study, so we did not need the Ethics Approval.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Hajizadeh N, Pourhoseingholi MA, Baghestani AR, et al. Bayesian adjustment for over-estimation and under-estimation of gastric cancer incidence across Iranian provinces. World J Gastrointest Oncol 2017;9:87-93. [Crossref] [PubMed]
- Munteanu A, Munteanu D, Tigan S, et al. How do surgical stress and low perioperative serum protein and albumin impact upon short term morbidity and mortality in gastric cancer surgery. Clujul Med 2017;90:71-85. [PubMed]
- Ma NF, Hu L, Fung JM, et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology 2008;47:503-10. [Crossref] [PubMed]
- Liu SS, Bai YS, Feng L, et al. Identification of CHD1L as an Important Regulator for Spermatogonial Stem Cell Survival and Self-Renewal. Stem Cells Int 2016;2016:4069543. [Crossref] [PubMed]
- Liu ZH, Zhang Q, Ding YJ, et al. Overexpression of CHD1L is associated with poor survival and aggressive tumor biology in esophageal carcinoma. Oncotarget 2017;8:74178-87. [Crossref] [PubMed]
- Xu X, He Y, Miao X, et al. Cell adhesion induces overexpression of chromodomain helicase/ATPase DNA binding protein 1-like gene (CHD1L) and contributes to cell adhesion-mediated drug resistance (CAM-DR) in multiple myeloma cells. Leuk Res 2016;47:54-62. [Crossref] [PubMed]
- Sun J, Zhang L, Zhao H, et al. CHD1L Regulates Cell Cycle, Apoptosis, and Migration in Glioma. Cell Mol Neurobiol 2016;36:565-76. [Crossref] [PubMed]
- Liu C, Fu X, Zhong Z, et al. CHD1L Expression Increases Tumor Progression and Acts as a Predictive Biomarker for Poor Prognosis in Pancreatic Cancer. Dig Dis Sci 2017;62:2376-85. [Crossref] [PubMed]
- Klein KM, Pendziwiat M, Eilam A, et al. The phenotypic spectrum of ARHGEF9 includes intellectual disability, focal epilepsy and febrile seizures. J Neurol 2017;264:1421-5. [Crossref] [PubMed]
- Striano P, Zara F. ARHGEF9 mutations cause a specific recognizable X-linked intellectual disability syndrome. Neurol Genet 2017;3:e159. [Crossref] [PubMed]
- Chen L, Hu L, Chan THM, et al. Abstract #3458: CHD1L suppresses the nucleus-to-mitochondria translocation of Nur77 to sustain hepatocellular carcinoma cell survival. Cancer Res 2009;69:3458.
- Ahn HM, Yoo JW, Lee S, et al. Peroxiredoxin 5 promotes the epithelial-mesenchymal transition in colon cancer. Biochem Biophys Res Commun 2017;487:580-6. [Crossref] [PubMed]
- Tsuda M, Cho K, Ooka M, et al. ALC1/CHD1L, a chromatin-remodeling enzyme, is required for efficient base excision repair. PLoS One 2017;12:e0188320. [Crossref] [PubMed]
- Liu M, Chen L, Ma NF, et al. CHD1L promotes lineage reversion of hepatocellular carcinoma through opening chromatin for key developmental transcription factors. Hepatology 2016;63:1544-59. [Crossref] [PubMed]
- Dou D, Zhao H, Li Z, et al. CHD1L Promotes Neuronal Differentiation in Human Embryonic Stem Cells by Upregulating PAX6. Stem Cells Dev 2017;26:1626-36. [Crossref] [PubMed]
- Yang M, Peng Y, Liu W, et al. Sirtuin 2 expression suppresses oxidative stress and senescence of nucleus pulposus cells through inhibition of the p53/p21 pathway. Biochem Biophys Res Commun 2019;513:616-22. [Crossref] [PubMed]
- Cheng W, Su Y, Xu F. CHD1L: a novel oncogene. Mol Cancer 2013;12:170. [Crossref] [PubMed]
- Dias AR, Azevedo BC, Alban LBV, et al. Gastric neuroendocrine tumor: review and update. Arq Bras Cir Dig 2017;30:150-4. [Crossref] [PubMed]
- Lu Y, Wei G, Liu L, et al. Direct targeting of MAPK8IP1 by miR-10a-5p is a major mechanism for gastric cancer metastasis. Oncol Lett 2017;13:1131-6. [Crossref] [PubMed]
- Mu QJ, Li HL, Yuan Y, et al. Chromodomain Helicase/ATPase DNA-Binding Protein 1-Like Gene (CHD1L) Expression and Implications for Invasion and Metastasis of Breast Cancer. PLoS One 2015;10:e0143030. [Crossref] [PubMed]
- Li Y, Chen L, Chan TH, et al. SPOCK1 is regulated by CHD1L and blocks apoptosis and promotes HCC cell invasiveness and metastasis in mice. Gastroenterology 2013;144:179-191.e4. [Crossref] [PubMed]
- Singh HR, Nardozza AP, Möller IR, et al. A Poly-ADP-Ribose Trigger Releases the Auto-Inhibition of a Chromatin Remodeling Oncogene. Mol Cell 2017;68:860-871.e7. [Crossref] [PubMed]
- Wang J, Liu M, Chen L, et al. Overexpression of N-terminal kinase like gene promotes tumorigenicity of hepatocellular carcinoma by regulating cell cycle progression and cell motility. Oncotarget 2015;6:1618-30. [Crossref] [PubMed]
- Tian F, Xu F, Zhang ZY, et al. Expression of CHD1L in bladder cancer and its influence on prognosis and survival. Tumour Biol 2013;34:3687-90. [Crossref] [PubMed]
- Ji X, Li J, Zhu L, et al. CHD1L promotes tumor progression and predicts survival in colorectal carcinoma. J Surg Res 2013;185:84-91. [Crossref] [PubMed]
- He WP, Zhou J, Cai MY, et al. CHD1L protein is overexpressed in human ovarian carcinomas and is a novel predictive biomarker for patients survival. BMC Cancer 2012;12:437. [Crossref] [PubMed]
- Su FR, Ding JH, Bo L, et al. Chromodomain helicase/ATPase DNA binding protein 1-like protein expression predicts poor prognosis in nasopharyngeal carcinoma. Exp Ther Med 2014;8:1745-50. [Crossref] [PubMed]
- Krepischi ACV, Maschietto M, Ferreira EN, et al. Genomic imbalances pinpoint potential oncogenes and tumor suppressors in Wilms tumors. Mol Cytogenet 2016;9:20. [Crossref] [PubMed]
- Chen L, Yuan YF, Li Y, et al. Clinical significance of CHD1L in hepatocellular carcinoma and therapeutic potentials of virus-mediated CHD1L depletion. Gut 2011;60:534-43. [Crossref] [PubMed]
- Ahel D, Horejsí Z, Wiechens N, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009;325:1240-3. [Crossref] [PubMed]
- Su Z, Zhao J, Xian G, et al. CHD1L is a novel independent prognostic factor for gastric cancer. Clin Transl Oncol 2014;16:702-7. [Crossref] [PubMed]
- Chen L, Hu L, Chan TH, et al. Chromodomain helicase/adenosine triphosphatase DNA binding protein 1-like (CHD1l) gene suppresses the nucleus-to-mitochondria translocation of nur77 to sustain hepatocellular carcinoma cell survival. Hepatology 2009;50:122-9. [Crossref] [PubMed]
- He LR, Ma NF, Chen JW, et al. Overexpression of CHD1L is positively associated with metastasis of lung adenocarcinoma and predicts patients poor survival. Oncotarget 2015;6:31181-90. [Crossref] [PubMed]
- Wu J, Zong Y, Fei X, et al. Presence of CHD1L over-expression is associated with aggressive tumor biology and is a novel prognostic biomarker for patient survival in human breast cancer. PLoS One 2014;9:e98673. [Crossref] [PubMed]
- Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol 2017;11:28-39. [Crossref] [PubMed]
- Wang R, Sun Y, Yu W, et al. Downregulation of miRNA-214 in cancer-associated fibroblasts contributes to migration and invasion of gastric cancer cells through targeting FGF9 and inducing EMT. J Exp Clin Cancer Res 2019;38:20. [Crossref] [PubMed]
- Xue Y, Zhang L, Zhu Y, et al. Regulation of Proliferation and Epithelial-to-Mesenchymal Transition (EMT) of Gastric Cancer by ZEB1 via Modulating Wnt5a and Related Mechanisms. Med Sci Monit 2019;25:1663-70. [Crossref] [PubMed]
- Alber M, Kalscheuer VM, Marco E, et al. ARHGEF9 disease: Phenotype clarification and genotype-phenotype correlation. Neurol Genet 2017;3:e148. [Crossref] [PubMed]
- Smith R, Sellou H, Chapuis C, et al. CHD3 and CHD4 recruitment and chromatin remodeling activity at DNA breaks is promoted by early poly (ADP-ribose)-dependent chromatin relaxation. Nucleic Acids Res 2018;46:6087-98. [Crossref] [PubMed]


