
Non-target genetic manipulation induces rhabdomyosarcoma in KrasPten-driven mouse model of ovarian cancer
Introduction
Ovarian cancer is a lethal gynecologic malignancy with the highest case-fatality rate (1), and it demonstrates five major histological subtypes (serous, endometrioid, mucinous, clear cell and transitional adenocarcinomas) (2). Ovarian surface epithelium (OSE), a single layer of cells covering the ovary, is the potential site of the origin of ovarian cancer. The “incessant ovulation” theory proposes that frequent ovulation and surface repair contribute to the malignant transformation (3). In the past few decades, significant progress has been made in uncovering insights into genetics of tumorigenesis in ovarian cancer. Indeed, distinct genetic alterations, coupled with other etiological factors, contribute to the development of different subtypes of ovarian tumor (4). Frequent somatic mutations in Kras and Pten discovered in ovarian endometrioid adenocarcinomas (OEAs) indicate that they play key roles in the etiology of this subtype (5,6).
Kras, a member of the Ras gene family, is involved in transmitting signals within cells (7). Specific alterations in the Kras gene can convert it into an active oncogene, which has been observed with varying frequencies in a significant proportion of human malignancies, including colorectal cancer, lung cancer, and pancreatic cancer (8-11). Homozygous deletion of Pten, a tumor suppressor gene, is also detected in a variety of human malignancies (12,13). Germ-line Pten mutations are frequently seen in patients with autosomal dominant cancer-predisposition syndromes (14), while somatic Pten mutations are detected in many tumors, including glioblastoma, OEAs, prostate cancer, and melanoma (15-18). A mouse model for OEAs has been previously generated through activation of Kras and deletion of Pten in transgenic mice using the Cre/lox recombination (19). The OEAs developed in mice exhibit histomorphology and biological behavior similar to that of human OEAs; therefore, this mouse model provides a useful tool for studies on ovarian cancer.
In order to generate the mouse model of OEAs driven by Kras activation and Pten deletion, Cre recombinase-expressing adenovirus (AdCre) were delivered into the ovarian bursa for activation of conditional alleles within the OSE. Subcutaneous lumps developed accidentally around the surgical incision post-AdCre administration. Cre-mediated recombination of Kras and Pten was verified in tumor tissues, and the activation of the downstream pathways was also consistent with specific genetic events. Furthermore, histopathological analysis revealed that these newly developed tumors were rhabdomyosarcoma (RMS). We verified that the leakage of adenovirus transformed healthy adjacent tissues into RMS. These findings imply that it is essential to avoid accidental non-target genetic manipulations to stably establish this mouse model of ovarian cancer. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2561).
Methods
Reagents and antibodies
The following reagents were used: Tris, glycine, sodium dodecyl sulfide and bovine serum albumin (BSA) (Sigma). Antibodies included: p-MAPKThr202/Tyr204, p-AKTSer473, p-mTORSer2448, p-S6KThr389 and p-4E-BP1Thr37746 (Cell signaling Technology); β-actin and myogenin (Genetex); myoD1 (Proteintech); vimentin, cytokeratin 8, Ki-67 and α-SMA (Abcam) and desmin (Servicebio Co. Ltd., China). Experiments were performed under a project license (NO.: L201501027) granted by ethics board of Sun Yat-sen University, in compliance with all national and/or institutional guidelines for the care and use of animals.
Generation of double transgenic KrasG12DPtenFl/Fl mice
LSL-KrasG12D (B6.129S4-Krastm4Tyj/J) and PtenFl/Fl (C;129S4-Ptentm1Hwu/J) mice were purchased from the Jackson Laboratory (Bar Harbor, USA). LSL-KrasG12D mice were crossed with PtenFl/Fl to generate pups harboring LSL-KrasG12DPtenFl/Fl (n=10), LSL-KrasG12D (n=10), and PtenFl/Fl (n=10). Eight-to-10-week-old female littermates were included in this study. DNA was extracted from the tail tissues for genotyping of mice following protocols provided on the Jackson Laboratory website (www.jax.org).
Administration of adenovirus
The recombinant adenovirus (AdCre-eGFP) was purchased from the Gene Transfer Vector Core, University of Iowa. We reconstituted the transduction system as described in a previous study (19). After using 5 units (U) of pregnant mare serum gonadotropin (Sigma) for 2 days, 5 U of human chorionic gonadotropin (Sigma) were used to promote synchronized ovulation, and 1.5 days later the animals were anesthetized and a dorsal incision was made to expose the ovaries. AdCre-eGFP was then delivered into the ovarian bursa via microinjection under a dissecting microscope. Then, we gently closed the dorsal incision by successively suturing the fascia, the muscle, and the skin with surgical stitches. Mice with tumors were sacrificed when they were moribund.
Fluorescence studies
Five days after the intrabursal administration of AdCre-eGFP, the treated-ovaries were harvested and snap-frozen using OCT (Sakura Fineteck). Seven-micrometer-thick cryo-sections of tumor tissues were prepared using cryo-ultramicrotome (Thermo). Sections were fixed in ice cold acetone for 15 minutes and the nuclei were stained with DAPI. Pictures were captured using a fluorescence microscope.
Recombination efficiency analysis
Genomic DNA was extracted from tumor and normal tissues using a TIANamp Genomic DNA Kit (TIANGEN, DP304), according to the manufacturer’s instructions. PCR was performed with primers designed for Cre-mediated recombination of Kras and Pten, followed by agarose gel electrophoresis of the PCR products. The primer sets used were as follows: KrasG12D (forward: GTCTGGAATTCCGCAAGCTA; reverse: GCACGCAGACTGTAGAGCAG), recombined KrasG12D (forward: GTCTTTCCCCAGCACAGTGC; reverse: CTCTTGCCTACGCCACCAGCTC), Pten (forward: CAAGCACTCTGCGAACTGAG; reverse: AAGTTTTTGAAGGCAAGATGC), and recombined Pten (forward: ACTCAAGGCAGGGATGAGC; reverse: GCTTGATATCGAATTCCTGCAGC). Transgenic alleles after genetic manipulation were as follows: (I) the Kras gene was heterozygous, expressing mutant KrasG12D and wild-type Kras respectively, (II) the Pten gene was homozygous; hence, showed complete deletion.
Histological analysis and immunohistochemistry (IHC)
Mouse tissues were excised immediately after euthanasia and fixed in 4% paraformaldehyde, followed by paraffin-embedding. Paraffin sections were deparaffinized, rehydrated, and stained using hematoxylin and eosin (H&E) or processed for IHC. For IHC, tissues sections were incubated with primary antibodies after antigen retrieval, and the immunostaining intensity was scored as previously described (20).
Western blotting
Tissues were homogenized and then lysed in RIPA buffer with protease inhibitors (Roche) and phosphatase inhibitors (Sigma). The lysates were loaded onto polyacrylamide gels and then transferred onto polyvinylidene difluoride (PVDF) membranes. Blots were blocked with 5% BSA in Tris-buffered saline-Tween 0.1% for 1 hour at room temperature; then, probed with antibodies overnight at 4 °C. Following a 1-hour incubation with the HRP-conjugated secondary antibody (Cell Signaling Technology) at 37 °C, detection was performed using a Pierce ECL Western Blotting Substrate system.
Statistical analysis
Unpaired, two-tailed Student’s t-tests were used for all analyses comparing two experimental groups. All data were exported and calculated into Microsoft Excel, and statistical analyses were performed using the GraphPad Prism software. A P value of <0.05 was considered statistically significant.
Results
Subcutaneous lumps developed accidentally in mouse model of ovarian cancer driven by oncogenic activation of Kras and deletion of Pten
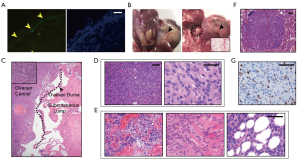
To achieve Cre-mediated recombination of Kras and Pten genes, adenoviruses expressing Cre recombinase and green fluorescent protein (AdCre-eGFP) were delivered into ovarian bursa after surgical exposure. This method was effective, as shown by the abundant GFP in the infected ovarian surface epithelial cells (OSECs) (Figure 1A). Then, we monitored the health of the animals every 3 days. Unexpectedly, large subcutaneous lumps rapidly developed in situ around the dorsal incision site in most of the compound mutant mice carrying LSL-KrasG12D and PtenFl/Fl (8/10, 80%), but not in mice carrying LSL-KrasG12D (0/10, 0%) or PtenFl/Fl (0/10, 0%) alone. These lumps had a shorter disease latency (range, 2–3.5 weeks) post-AdCre-eGFP administration before the mice exhibited the signs of ovarian cancer. More importantly, the rapid and infiltrative growth of these lumps complicated the ovarian cancer progression because of disordered symptoms. The animals with the subcutaneous lumps were moribund approximately 5 weeks post-AdCre-eGFP administration (range, 4.5–6.5 weeks), probably because of the end-stage cachexia and multiple organ dysfunction syndrome under high tumor burden. At the same time, the remaining two mice without lumps were healthy with palpable ovarian tumor. We euthanized the moribund animals and dissected the lumps anatomically. The lump appeared separate from the OEAs, and appeared to originate from the abdominal wall (Figure 1B). Furthermore, the lumps were circular (diameter between 2.5 to 6.2 cm) and displayed a firm, white and whorled appearance in sections (Figure 1B). H&E staining revealed that the lumps were separate from the ovarian cancer, and demonstrated remarkably different histomorphology from the typical glandular epithelium of OEAs (Figure 1C). Particularly, the lumps exhibited increased cellularity and frequent mitotic figures, and were made of spindle cells in sweeping fascicles (Figure 1D). Furthermore, frequent focal necrosis (Figure 1E, left), invasion of adjacent skeletal muscle (Figure 1E, middle), and adipose tissue invasion (Figure 1E, right) were observed in most cases. Some mice with lumps developed lung metastases (2/8, 25%; Figure 1F). Consistent with the aggressive phenotype, >30% of cells were positive for Ki-67 in the nuclei (Figure 1G). Overall, these results demonstrated that subcutaneous lumps with a malignant phenotype developed in the transgenic mice because of Kras activation and Pten deletion.
Kras activation and Pten deletion are crucial for the development of the subcutaneous lumps
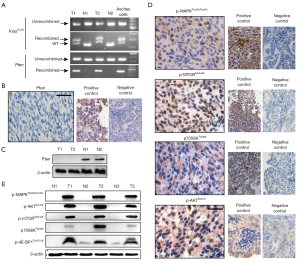
To understand the molecular mechanism underlying the efficient development of these subcutaneous lumps, the specific genetic events caused by Kras activation and Pten deletion were explored. We used PCR analysis to identify the recombination of the conditional alleles in the tumor, with the normal tissues from the corresponding mice as a control (Figure 2A). Cre-mediated recombination was further confirmed by examining Pten protein expression in the tumor using IHC staining (Figure 2B) and Western blotting (Figure 2C). We reasoned that the activation of the pathways downstream oncogenic Kras activation and Pten deletion contributed to the pathogenesis of these lumps. As expected, high levels of phosphorylated MAPK, an effector of oncogenic Kras, suggested the activation of MAPK pathway (Figure 2D). Furthermore, enhanced phosphorylation of AKT, mTOR, and p70S6K indicated the activation of the PI3K-Akt-mTOR pathway (Figure 2D). We further examined similar changes in the levels of key proteins in the tumor mass preparations using Western blotting (Figure 2E). Collectively, these findings indicated that oncogenic activation of Kras and deletion of Pten were crucial in the pathogenesis of the subcutaneous tumor.
Histopathological analysis identified the lumps as aggressive soft tissue sarcomas (STSs) with a mesenchymal origin
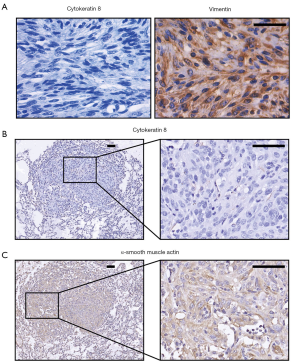
We first evaluated the histopathological signatures of the subcutaneous tumors to understand the type of disease to which they belong. IHC revealed that the lumps did not show epithelial differentiation, indicated by negative staining of cytokeratin 8 (Figure 3A). Moreover, the intense positive staining for vimentin, the canonical mesenchymal marker, implied the mesenchymal origin of the tumor (Figure 3A). Consistently, the lung metastases showed no staining for cytokeratin 8 but were positively stained for α-smooth muscle actin (α-SMA) (Figure 3B,C). Thus, we concluded that the subcutaneous lumps developed in our model of ovarian cancer were mesenchymal-derived STSs.
The STSs showed properties of RMS
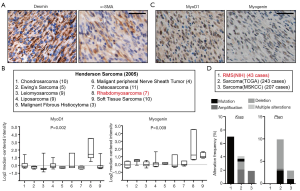
AS STSs have histological diversity, intricate molecular profiles, various biological behaviors, and different clinical syndromes, we next explored the specific histopathological properties of the STSs to make a differential diagnosis. According to the diagnostic standard in human STSs, diffuse expression of desmin and focal positivity for α-SMA in the subcutaneous lumps indicated them to be RMS, leiomyosarcoma, or low-grade myofibroblastic sarcoma (Figure 4A). We analyze published transcriptional data of diverse kind of human sarcomas from the Oncomine database (http://www.oncomine.org) and found that the up-regulation of myoD1 and myogenin were specific to RMS (Figure 4B). Indeed, the lumps showed specific and intensely positive staining for myogenic markers, myoD1 and myogenin (Figure 4C), which are sensitive and specific diagnostic molecular biomarkers of RMS (21). Based on these results, we diagnosis the STSs developed in our model as RMS. To reveal genomic alterations of Kras and Pten in human RMS, we then analyzed their alterations at genomic levels using cBioPortal database (https://www.cbioportal.org/). Kras mutations were found in 6.9% RMS while Pten loss is genomically rare, though it can be found in other types of sarcomas (Figure 4D). Collectively, these results indicated that the lumps developed in our model were RMSs.
Reduced leakage of AdCre-eGFP decreased the formation of the subcutaneous lumps
The anatomical location and course of the lumps were different from ovarian cancer; therefore, we hypothesized that the accidental exposure of the adjacent tissues to AdCre-eGFP is responsible for the lump development. To improve our mouse model work great, we replicated the genetic manipulations while modifying several methods to significantly decrease the incidence rate of the subcutaneous lumps. First, the syringes we initially used were too large to easily control the volume injected, which frequently resulted in injecting >5 µL of the Adcre-eGFP solution, which is the maximum volume recommended in the previous reports (19,22,23). Second, the medium containing AdCre-eGFP was transparent, making it difficult to track the intrabursal injection and to ensure that there is no spillage or leakage. Thus, we added trypan blue to the medium for tracking the spread of the injected solution. Additionally, we were not sufficiently skillful to perform the intrabursal injection in our early attempts, which likely increased the chance for AdCre-eGFP leakage. In addition, we also performed a peritoneal lavage with saline to decrease the accidental exposure of AdCre-eGFP to other normal tissues. With the above improvements to our genetic manipulation protocols, the incidence of RMS was significantly decreased (1/10, 10%). This makes our mouse model as a practical tool for investigations of ovarian cancer.
Discussion
In this study, we demonstrated that subcutaneous lumps unexpectedly develop around the surgical incision during the establishment of a mouse model of ovarian cancer driven by Kras activation and Pten deletion. Cre-mediated recombination of Kras and Pten were confirmed in the tumor, and the activation of downstream pathways was consistent with the specific genetic events. Histological and histopathological analysis determined these subcutaneous tumors as RMSs. We confirmed that the malignant transformation of non-target tissues due to the accidental exposure to adenovirus contributed to the RMS development.
Activation of oncogenes and/or the loss of tumor suppressing genes causes alteration of healthy tissues into malignancies. Tissue-specific context, that is, the cells of origin of tumors, have been considered as important determinants of the susceptibility to oncogenic transformation (24,25). Herein, transformation of non-specific cell types can occur upon in vivo genetic manipulations during establishment of the mouse model, which has been indicated in previous reports. Low specificity of cell/tissue-specific promoters in these genetically modified mouse models partially explains the non-specific genetic alterations (26-29). However, in these models that are dependent on genetic manipulations by virtue of adenovirus or lentivirus infections, direct exposure of adjacent tissues and/or the accidental exposure due to indefinite borderlines contribute to such unexpected results. According to previous reports, AdCre-mediated conditional mutations in Kras and p53 in skeletal muscle tissues induce high-grade sarcomas with myofibroblastic differentiation (30), whereas dual Pten and p53 suppression in the smooth muscle lineage promotes the progression of sarcoma (31). In addition, simultaneous Kras activation and p53 inactivation induces sarcoma formation in the soft tissues due to accidental leakage of recombinant AdCre from the suture site; Further, it leads to urothelial hyperplasia in the bladder (32). In an effort to establish ovarian cancer mouse model, simultaneous inactivation of p53 and Rb1 in OSECs induced epithelial ovarian cancer (33,34), whereas conditional inactivation of Brca1, p53, and Rb by intrabursal administration of AdCre unexpectedly transforms the bursal membrane cells to develop leiomyosarcomas (23). In our study, the exposure of OSECs and abdominal muscle tissues to AdCre-eGFP induced ovarian cancer and RMS, respectively, in mice harboring conditional alleles of LSL-KrasG12D and PtenFl/Fl. We postulated that the leakage from the pin hole was mainly responsible for the accidental development of RMS. Development of RMS was remarkably reduced after improvement in technique, which facilitated this mouse model to be used as a practical model for investigations of ovarian cancer.
RMS is an aggressive subtype of STSs with vague etiology and vast heterogeneity (35). Although it is thought to arise from a skeletal muscle lineage, RMS is a malignant tumor with mesenchymal origin (36). Owing to the diversity of clinicopathologic characteristics, morphologic patterns, and histologic categories, molecular markers identified using IHC, such as desmin, α-SMA, myoD1 and myogenin, are considered an important tool for the positive diagnosis of STSs (37). Importantly, myogenin is a specific and an independent marker of poor survival for patients with RMS (38-40). From a genomic perspective, 3.9% of human RMS samples carry Pten mutations, whereas 0% to 44% samples carry Ras mutations, suggesting the haploinsufficiency of Ras or Pten mutations alone. Nevertheless, the coordinated genetic alterations of Kras and Pten in RMS are poorly unknown. Recently, integrated genetic and epigenetic analyses of human RMS have unveiled the prevalence of the receptor tyrosine kinase/Ras/Pik3ca axis and Pten methylation, which are reported in up to 93% and 70% of cases, respectively (41). Therefore, simultaneous Kras activation and Pten inactivation may be essential for the development of RMS, which complicates the establishment of a stable model of OEA in mice harboring conditional alleles of LSL-KrasG12D and PtenFl/Fl.
In conclusion, non-specific genetic alterations induce development of RMS in the mouse model of KrasPten-driven ovarian cancer, indicating the importance of carefully avoiding inappropriate exposure of genetic manipulations to non-specific tissues.
Acknowledgments
We thank Dr. Daniela Dinulescu of the Department of Pathology at BWH (Brigham and Women’s Hospital) for technical support with surgical skills.
Funding: This work was funded by grants from
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist Available at http://dx.doi.org/10.21037/tcr-20-2561
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2561
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2561
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2561). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (NO.: L201501027) granted by ethics board of Sun Yat-sen University, in compliance with all national and/or institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Cho KR, Shih IeM. Ovarian cancer. Annu Rev Pathol 2009;4:287-313. [Crossref] [PubMed]
- Kurman RJ, Shih Ie M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol 2010;34:433-43. [Crossref] [PubMed]
- Lengyel E. Ovarian cancer development and metastasis. Am J Pathol 2010;177:1053-64. [Crossref] [PubMed]
- Stewart CJ, Walsh MD, Budgeon CA, et al. Immunophenotypic analysis of ovarian endometrioid adenocarcinoma: correlation with KRAS mutation and the presence of endometriosis. Pathology 2013;45:559-66. [Crossref] [PubMed]
- Tanwar PS, Kaneko-Tarui T, Lee HJ, et al. PTEN loss and HOXA10 expression are associated with ovarian endometrioid adenocarcinoma differentiation and progression. Carcinogenesis 2013;34:893-901. [Crossref] [PubMed]
- Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol 2003;4:373-84. [Crossref] [PubMed]
- Jancik S, Drabek J, Radzioch D, et al. Clinical relevance of KRAS in human cancers. J Biomed Biotechnol 2010;2010:150960. [Crossref] [PubMed]
- Phipps AI, Buchanan DD, Makar KW, et al. KRAS-mutation status in relation to colorectal cancer survival: the joint impact of correlated tumour markers. Br J Cancer 2013;108:1757-64. [Crossref] [PubMed]
- Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc 2009;6:201-5. [Crossref] [PubMed]
- Eser S, Schnieke A, Schneider G, et al. Oncogenic KRAS signalling in pancreatic cancer. Br J Cancer 2014;111:817-22. [Crossref] [PubMed]
- Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol 1999;9:125-8. [Crossref] [PubMed]
- Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 2009;4:127-50. [Crossref] [PubMed]
- Marsh DJ, Dahia PL, Zheng Z, et al. Germline mutations in PTEN are present in Bannayan-Zonana syndrome. Nat Genet 1997;16:333-4. [Crossref] [PubMed]
- Wang SI, Puc J, Li J, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res 1997;57:4183-6. [PubMed]
- Storey DJ, Rush R, Stewart M, et al. Endometrioid epithelial ovarian cancer: 20 years of prospectively collected data from a single center. Cancer 2008;112:2211-20. [Crossref] [PubMed]
- Deocampo ND, Huang H, Tindall DJ. The role of PTEN in the progression and survival of prostate cancer. Minerva Endocrinol 2003;28:145-53. [PubMed]
- Wu H, Goel V, Haluska FG. PTEN signaling pathways in melanoma. Oncogene 2003;22:3113-22. [Crossref] [PubMed]
- Dinulescu DM, Ince TA, Quade BJ, et al. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med 2005;11:63-70. [Crossref] [PubMed]
- Zhao X, Fang Y, Yang Y, et al. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy 2015;11:1849-63. [Crossref] [PubMed]
- Sebire NJ, Malone M. Myogenin and MyoD1 expression in paediatric rhabdomyosarcomas. J Clin Pathol 2003;56:412-6. [Crossref] [PubMed]
- Wu R, Hendrix-Lucas N, Kuick R, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer Cell 2007;11:321-33. [Crossref] [PubMed]
- Clark-Knowles KV, Senterman MK, Collins O, et al. Conditional inactivation of Brca1, p53 and Rb in mouse ovaries results in the development of leiomyosarcomas. PLoS One 2009;4:e8534. [Crossref] [PubMed]
- Schneider G, Schmidt-Supprian M, Rad R, et al. Tissue-specific tumorigenesis: context matters. Nat Rev Cancer 2017;17:239-53. [Crossref] [PubMed]
- Sharma A, Sen JM. Molecular basis for the tissue specificity of beta-catenin oncogenesis. Oncogene 2013;32:1901-9. [Crossref] [PubMed]
- Haruyama N, Cho A, Kulkarni AB. Overview: engineering transgenic constructs and mice. Curr Protoc Cell Biol 2009;Chapter 19:Unit-19.10.
- Lammel S, Steinberg EE, Foldy C, et al. Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron 2015;85:429-38. [Crossref] [PubMed]
- Elefteriou F, Yang X. Genetic mouse models for bone studies--strengths and limitations. Bone 2011;49:1242-54. [Crossref] [PubMed]
- Jeffery E, Berry R, Church CD, et al. Characterization of Cre recombinase models for the study of adipose tissue. Adipocyte 2014;3:206-11. [Crossref] [PubMed]
- Kirsch DG, Dinulescu DM, Miller JB, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med 2007;13:992-7. [Crossref] [PubMed]
- Guijarro MV, Dahiya S, Danielson LS, et al. Dual Pten/Tp53 suppression promotes sarcoma progression by activating Notch signaling. Am J Pathol 2013;182:2015-27. [Crossref] [PubMed]
- Yang X, La Rosa FG, Genova EE, et al. Simultaneous activation of Kras and inactivation of p53 induces soft tissue sarcoma and bladder urothelial hyperplasia. PLoS One 2013;8:e74809. [Crossref] [PubMed]
- Flesken-Nikitin A, Hwang CI, Cheng CY, et al. Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche. Nature 2013;495:241-5. [Crossref] [PubMed]
- Barger CJ, Zhang W, Hillman J, et al. Genetic determinants of FOXM1 overexpression in epithelial ovarian cancer and functional contribution to cell cycle progression. Oncotarget 2015;6:27613-27. [Crossref] [PubMed]
- Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer 2015;15:426-39. [Crossref] [PubMed]
- Dagher R, Helman L. Rhabdomyosarcoma: an overview. Oncologist 1999;4:34-44. [Crossref] [PubMed]
- Stock N, Chibon F, Binh MB, et al. Adult-type rhabdomyosarcoma: analysis of 57 cases with clinicopathologic description, identification of 3 morphologic patterns and prognosis. Am J Surg Pathol 2009;33:1850-9. [Crossref] [PubMed]
- Dias P, Chen B, Dilday B, et al. Strong immunostaining for myogenin in rhabdomyosarcoma is significantly associated with tumors of the alveolar subclass. Am J Pathol 2000;156:399-408. [Crossref] [PubMed]
- Kumar S, Perlman E, Harris CA, et al. Myogenin is a specific marker for rhabdomyosarcoma: an immunohistochemical study in paraffin-embedded tissues. Mod Pathol 2000;13:988-93. [Crossref] [PubMed]
- Heerema-McKenney A, Wijnaendts LC, Pulliam JF, et al. Diffuse myogenin expression by immunohistochemistry is an independent marker of poor survival in pediatric rhabdomyosarcoma: a tissue microarray study of 71 primary tumors including correlation with molecular phenotype. Am J Surg Pathol 2008;32:1513-22. [Crossref] [PubMed]
- Seki M, Nishimura R, Yoshida K, et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun 2015;6:7557. [Crossref] [PubMed]


