
AT-rich interactive domain1A determines sensitivity to oxaliplatin in gastric cancer cells
Introduction
Gastric cancer is considered as one of the most common malignant tumors with a high mortality, ranking the third among all cancer-related deaths in the world (1,2). Additionally, gastric cancer is a highly heterogeneous disease and its traditional histopathological classification is difficult to meet clinical needs. The Cancer Genome Atlas (TCGA) project divides gastric cancer into 4 subtypes: Epstein-Barr virus, microsatellite instability, and genomically stable and chromosomal instability (3). Therefore, it is particularly important to find out molecular markers related to the pathogenesis and prognosis of gastric cancer.
AT-rich interactive domain1A gene (ARID1A) is a crucial member of SWI/SNF chromatin-remodeling complexes (4). It has helicase and ATPase activities and plays a role in embryonic development, tissue growth, cell proliferation, differentiation and apoptosis, as well as regulations of DNA synthesis, gene transcription, and DNA damage repair (5,6). In recent years, studies have found that in addition to ovarian cancer and head and neck cancer, ARID1A mutations are also widely found in liver cancer, gastric cancer, lung cancer and kidney cancer, which have become a recent research hotspot (7-12). Among them, ARID1A mutation rate in gastric cancer tissues is 8–27%, and this gene mutation often leads to the loss or decrease of protein expression (13,14). Low expression of ARID1A gene is related with tumor progression, lymph node metastasis and vascular invasion (15,16), which is also one of the factors predicting poor prognosis of this tumor (17). Therefore, ARID1A gene mutation is likely to be a key factor in the gastric cancer development.
At present, the treatment of advanced gastric cancer is still based on chemotherapy. The first-line chemotherapy regimens recommended by the National Comprehensive Cancer Network guidelines for gastric cancer include oxaliplatin, platinum, taxanes, and fluorouracil. Oxaliplatin is a third-generation platinum antitumor drug with high efficiency and low toxicity (18,19). Therefore, the insensitivity or secondary drug resistance of oxaliplatin to gastric cancer is one of the vital causes of tumor progression. Lyu et al. have shown that ARID1A siRNA decreased the expression of ARID1A significantly, which led to cisplatin chemosensitivity and also cell apoptosis reduction in ES2 ovarian cancer cells via the regulation of AKT expression (20). For gastric cancer, there is no clinical or laboratory evidence to show whether the low expression of ARID1A affects the sensitivity of gastric cancer to oxaliplatin.
Therefore, this study established an ARID1A gene silencing model of gastric cancer cell transfected with short hairpin RNA (shRNA)-lentivirus to investigate the effects of ARID1A gene silencing on the biological behavior of gastric cancer cells and the sensitivity of gastric cancer cells to oxaliplatin after ARID1A gene silencing. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2384).
Methods
Cell culture
MGC-803 and AGS cells were obtained from the Key Laboratory of Digestive Tract Malignancies, Ministry of Education, Fujian Medical University (Fujian, China). All cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Hyclone, USA) supplemented with 10% fetal bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd. China), 100 U/mL penicillin and 100 mg/mL streptomycin (Hyclone, USA) in a humidified atmosphere of 5% CO2 at 37 °C. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study does not need ethics approval because of no animals or people involved.
shRNAs
After transfection for 48 h, the gastric cancer cell line was screened with puromycin, and the transfection was observed under an inverted fluorescence microscope. Furthermore, the cells in the negative empty vector control group (NC) were treated with empty vector. Design and synthesis of ARID1A gene lentiviral shRNAs sequences were shown in Table 1.
Table 1
| Gene | Primer sequence (5'-3') |
|---|---|
| shRNA | Sense: 5'-GCCCTGAACAATAACCTCACTTCAAGAGAGTGAGGTTATTGTTCAGGGTTTTTTACGCGTg-3' |
| Antisense: 5'-ACGCGTAAAAAACCCTGAACAATAACCTCACTCTCTTGAAGTGAGGTTATTGTTCAGGGCg-3' |
shRNA, short hairpin RNA.
Quantitative reverse-transcription PCR (qRT-PCR)
According to the manufacturer’s protocol, total RNA was extracted from MGC-803 and AGS cells using Trizol Reagent (Invitrogen, CA), then the isolated RNA was reverse-transcribed into cDNA via PrimeScriptTM RT reagent Kit with gDNA Eraser (TaKaRa, Japan). Real-time PCR amplification of the gene of interest was listed in Table 2. The mRNA levels were calculated using the 2-∆∆Ct method. The RNA expression values were normalized using β-actin mRNA level.
Table 2
| Gene | Primer |
|---|---|
| ARID1A-forward | 5-TTAACTCCAGCCACCAAAATGAAC-3 |
| ARID1A-reverse | 5-ATAGAGGCGATAGAGGTCCAGAGG-3 |
| GAPDH-forward | 5-CCTGTTCGACAGTCAGCCG-3 |
| GAPDH-reverse | 5' 5-CGACCAAATCCGTTGACTCC-3 |
ARID1A, AT-rich interactive domain1A gene.
Western blot analysis
The total proteins of MGC-803 and AGS cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Beyotime Institute of Biotechnology, China). The protein concentration was measured by the BCA protein assay kit (Beyotime Institute of Biotechnology, China); 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was firstly used to separate protein samples (50 µg), followed by transferring samples onto nitrocellulose membranes which were blocked with 5% non-fat milk at room temperature for 1 h. Subsequently, the membranes were incubated separately with rabbit anti-human ARID1A polyclonal antibody (1:1,000; Millipore, Bedford, MA, USA) and mouse anti-human β-actin monoclonal antibody (1:1,000; Xiamen Lulong Biotech Co., Ltd.) at 4 °C overnight, followed by 3 times washing with citrate buffer (TBST) containing 0.05% Tween 20 next day. Finally, the membranes were incubated at room temperature for 2 h using either a horseradish peroxide-conjugated mouse IgG secondary antibody (1:1,000; Abcam, UK) or rabbit IgG secondary antibody (1:5,000; Abcam, UK). After that, an enhanced chemiluminescence kit (ECL; Xiamen Lulong Biotech Co., Ltd., China) was used to visualize proteins and the densitometric analysis was conducted to quantify the protein band intensity using Image J image analysis software.
Cell Counting Kit-8 (CCK-8) assay
The MGC-803 and AGS cells in logarithmic growth phase were digested with 0.25% trypsin to prepare a single cell suspension, which was inoculated into 96-well plates at a density of 3×103 cells/well at 37 °C with 5% CO2. Ten µL of CCK-8 solution was then added to each well. A microplate reader was used to measure the absorbance value (OD value) at 450 and 630 nm after 1 h incubation. The proliferation activity of the cells was represented by the A value, which was proportional to the number of living cells.
Flow cytometry
As described in the instructions of Annexin V-PE/7-AAD apoptosis detection kit (BD Biosciences, USA), the MGC803 and AGS cells were separately digested, centrifuged and collected, then washed three times with cold phosphate buffer (PBS) to adjust the cell density to 2×105/mL. Binding buffer (200 µL) was added to resuspend the cells. Then 5 µL of Annexin V-PE staining solution was added in EP tubes, being mixed gently; followed by incubation for 15 min at 2–8 °C in the dark. Ten µL of 7-AAD staining solution was added to stain DNA for 5 min at 2–8 °C in the dark. The apoptotic rate was measured by flow cytometry (BD Biosciences, USA) within 1 h.
Transwell invasion assay
The upper chamber of the transwell chamber (Corning, USA) was precoated with 50 µL of Matrigel (1:8 dilution) and removed to a 37 °C incubator for 2 h. While 600 µL of complete medium was added to the lower chamber, 200 µL cell suspension (1×105 cells/mL) was added to the upper chamber and incubated for 12 h. The membrane was washed with PBS. The cells were wiped in the chamber, and then fixed with 4% paraformaldehyde for 20 min. After removing the non-migrated cells with cotton swabs, the migrating cells were stained with crystal violet for 20 min. Fields were selected at random to count and photograph cells under a light microscope. Cells passed through the Matrigel were analyzed to estimate the invasion ability. Cell invasion assay was detected by a transwell chamber invasion model (Corning, USA).
Statistical analysis
SPSS software version 20.0 was used to perform statistical analyses and the experimental measurement data were present as mean ± SD. The comparison of mean between the two groups was conducted by independent sample t test. Comparisons between groups were analyzed using one-way analysis of variance (one-way ANOVA). Differences were considered as significant when P<0.05.
Results
AGS and MGC-803 cells express ARID1A at both transcriptional and translational levels
To detect the mRNA and protein expression levels of ARID1A in gastric cancer cells (AGS and MGC-803 cells), qRT-PCR and Western blot were performed. As can be seen from Figure S1A,S1B, AGS and MGC-803 cells expressed ARID1A at both transcriptional and translational levels. Therefore, we made the follow-up test on the AGS and MGC-803 cells.
Transfection efficiency and protein expression of ARID1A gene silencing
To screen stable gastric cancer cells with ARID1A gene silencing, shRNA lentivirus was transfected into gastric cancer cells, and the transfection was observed under an inverted fluorescence microscope. We found that the transfection efficiency of green fluorescence was over 80% after the transfection of AGS and MGC-803 cells for 72 h with MOI value of 20 (Figure 1A,B,C,D). Subsequently, the results from Western blot indicated that the protein expression levels of shARID1A in AGS and MGC-803 cells were significantly decreased (P<0.001) (Figure 1E,F,G,H). Therefore, the lentivirus carrying a specific shRNA sequence could be selected for transfection into gastric cancer cells and used in subsequent experiments.
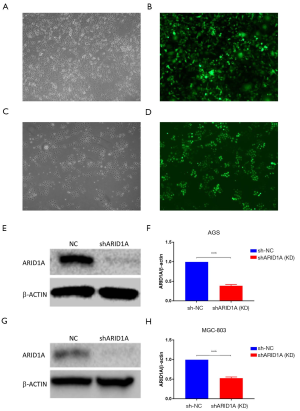
Effect of ARID1A gene silencing on gastric cancer cells
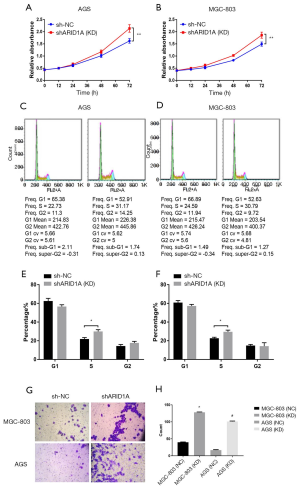
To discover the impact of ARID1A gene silencing on gastric cancer cells, shRNA lentivirus was transfected into AGS and MGC-803 cells and detected by CCK-8. It was found that ARID1A gene silencing significantly enhanced cell proliferation activity (P<0.01) (Figure 2A,B). The results of flow cytometry showed that the ARID1A gene silencing remarkedly increased the number of AGS and MGC-803 cells in S phase (Figure 2C,D,E,F), while decreased in G1 phase (P<0.05) (Figure 2C,D,F), indicating that ARID1A gene silencing could promote gastric cancer cells into the cell cycle. Invasive ability of gastric cancer cells was assayed in transwell chambers, the results indicated that the number of invading cells of gastric cancer after ARID1A gene silencing was significantly increased (P<0.05) (Figure 2G,H), suggesting that ARID1A gene silencing could enhance the invasion ability of gastric cancer cells. The above findings indicated that ARID1A gene silencing could enhance the activity of gastric cancer cells proliferation, promote their entry into the cell cycle, and improve their invasive ability.
Silencing ARID1A gene reduces the sensitivity of gastric cancer cells to oxaliplatin
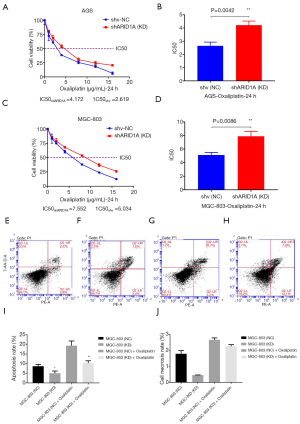
To further investigate the effect of ARID1A gene silencing on the sensitivity of gastric cancer cells to oxaliplatin, AGS and MGC-803 cells were treated with oxaliplatin at concentrations of 1, 2, 4, 8, 12 and 16 µg/mL and then subjected to CCK-8. The results clarified that ARID1A gene silencing significantly reduced the inhibition rate of oxaliplatin on gastric cancer cells in a concentration-dependent manner, and the IC50 increased significantly (P<0.01) (Figure 3A,B,C,D). Then, the apoptosis rate of gastric cancer cells treated with 10 µg/mL oxaliplatin was detected using flow cytometry. Furthermore, the result also indicated that ARID1A gene silencing reduced the apoptosis rate of gastric cancer cells significantly (P<0.05) (Figure 3E,F,G,H,I,J), suggesting that ARID1A gene silencing could reduce the sensitivity of gastric cancer cells to oxaliplatin.
Discussion
The most notable finding of this study was that ARID1A gene silencing is a promoting factor in gastric cancer cells and can reduce the sensitivity of gastric cancer cells to oxaliplatin. This finding further explores the molecular markers associated with the pathogenesis of gastric cancer. The occurrence and development of gastric cancer are complex processes influenced by multiple factors (21). Therefore, it is particularly important to find molecular markers related to the gastric cancer pathogenesis, which provide a basis for the exploration of targeted drugs for individualized treatment of gastric cancer. In the whole genome exon sequencing of gastric cancer tissues, genes with higher mutation rate in gastric cancer are TP53, PIK3CA, ARID1A (22). It is reported in the literature that ARID1A frequently undergoes gene mutations in various tumors, suggesting that it may be a potential tumor suppressor gene (23,24). Oxaliplatin based chemotherapy plays a significant role in the treatment of gastric cancer, but many cancer patients are resistant to platinum drugs, which seriously reduces the efficacy of platinum drugs and limits their clinical application (25). The high ARID1A mutation rate in gastric cancer might be the reason for the platinum resistance. Besides, a previous study about Jurkat leukemia cells showed that the FAS-mediated apoptosis was decreased after knocking out the ARID1A gene (26). In the current study, we found that ARID1A gene silencing could increase the proliferation activity of gastric cancer cells, reduce apoptosis, promote its entry into the cell cycle, and improve cell invasion. It was also proved that the mutation of ARID1A gene was related with the drug resistance of gastric cancer cells.
The AGS and MGC-803 cells are typical gastric cancer cells (27). Zhao et al. (28) have found that ARID1A inactivation causes defects in telomere cohesion, which selectively eliminated gross chromosome aberrations during mitosis. Colony formation capability of single cells in G2/M, but not G1 phase, was significantly reduced by ARID1A inactivation. Williamson et al. (29) also have point out that ARID1A defective cells have a delayed progression through the cell cycle and specifically delayed progression through S and G2/M, in ARID1A mutant tumour cells, inhibition of ATR triggers premature mitotic entry, genomic instability and apoptosis. The results of Williamson et al. were similar to our study, either MGC803 or AGS cell lines, ARID1A gene silencing led to the cell cycle delayed progression, and there were significant differences in S phase (P<0.05). In this study, we illustrated the biological effects of ARID1A gene silencing on gastric cancer cells and the effect on the sensitivity of gastric cancer cells to oxaliplatin. Besides, ARID1A gene silencing could also reduce the sensitivity of gastric cancer cells to oxaliplatin.
Oxaliplatin has a good effect on the treatment of colon cancer, lung cancer and gastric cancer, and is frequently used in clinical applications (18,30). However, many patients with gastric cancer are clinically insensitive or secondary to oxaliplatin, leading to tumor progression. The study results showed that ARID1A gene silencing promoted the proliferation and invasion of gastric cancer cells, and affected the biological behavior of gastric cancer cells. Lyu et al. (20) silenced the ARID1A gene by siRNA technology, and found that the sensitivity of ES2 ovarian cancer cell line to cisplatin was decreased, indicating that the resistance to cisplatin in ovarian cancer might be related to the low expression of ARID1A, which was consistent with our results.
Yet, the drug resistance mechanism of gastric cancer cells induced by ARID1A gene silencing is still unclear. We speculate that the mechanism was related to Akt signaling pathway. Akt, or protein kinase B, plays an important part in cell proliferation and apoptosis (31). Studies have shown that loss of nuclear ARID1A expression is associated with increased AKT phosphorylation in clear ovarian cell carcinoma tumors (32). In addition, the deletion of ARID1A promotes the proliferation of cancer cells by activating the AKT signaling pathway in GC cell lines (33). ARID1A gene silencing controls cell proliferation and apoptosis by promoting the phosphorylation of Akt, thereby reducing the sensitivity of cells to oxaliplatin. Next, we will run Western blotting for p-AKT in both the cell lines to find out whether increased p-AKT might be the mechanism for increased proliferation and/or cell motility after depletion of ARID1A. And the way in which ARID1A gene silencing affects the phosphorylation level of Akt requires further research.
Conclusions
In summary, this study confirms that ARID1A is a potential tumor suppressor gene, and we verified for the first time that ARID1A gene silencing can reduce the sensitivity of gastric cancer cells to oxaliplatin, providing a theoretical basis for predicting the efficacy of oxaliplatin-based chemotherapy in patients with gastric cancer and formulating individualized treatment plans.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2384
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2384
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2384
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2384). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The present study does not need ethics approval because of no animals or people involved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kanda M, Kodera Y. Recent advances in the molecular diagnostics of gastric cancer. World J Gastroenterol 2015;21:9838-52. [Crossref] [PubMed]
- Price TJ, Shapiro JD, Eva S, et al. Management of advanced gastric cancer. Expert Rev Gastroenterol Hepatol 2012;6:199-208. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [Crossref] [PubMed]
- Dallas PB, Pacchione S, Wilsker D, et al. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol 2000;20:3137-46. [Crossref] [PubMed]
- Flores-Alcantar A, Gonzalez-Sandoval A, Escalante-Alcalde D, et al. Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res 2011;345:137-48. [Crossref] [PubMed]
- Wang DD, Chen Y, Pan K, et al. Decreased Expression of the ARID1A Gene Is Associated with Poor Prognosis in Primary Gastric Cancer. PLoS One 2012;7:e40364. [Crossref] [PubMed]
- Takeda T, Kouji B, Ryuichiro O, et al. ARID1A gene mutation in ovarian and endometrial cancers Oncol Rep 2016;35:607-13. (Review). [Crossref] [PubMed]
- Wu RC, Wang TL, Shih IeM. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther 2014;15:655-64. [Crossref] [PubMed]
- Mao TL, Shih IeM. The roles of ARID1A in gynecologic cancer. J Gynecol Oncol 2013;24:376-81. [Crossref] [PubMed]
- Guan B, Wang TL, Shih IeM. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodeling, is a tumor suppressor in gynecologic cancers. Cancer Res 2011;71:6718-27. [Crossref] [PubMed]
- Wu JN, Roberts CW. ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor? Cancer Discov 2013;3:35-43. [Crossref] [PubMed]
- Guo XQ, Zhang QX, Huang WR, et al. Tumor suppressor role of chromatin-remodeling factor ARID1A. Yi Chuan 2013;35:255-61. [Crossref] [PubMed]
- Cheng CW, Licence D, Cook E, et al. Activation of mutated K-ras in donor endometrial epithelium and stroma promotes lesion growth in an intact immunocompetent murine model of endometriosis. J Pathol 2011;224:261-9. [Crossref] [PubMed]
- Nyiraneza C, Marbaix E, Smets M, et al. High risk for neoplastic transformation of endometriosis in a carrier of lynch syndrome. Fam Cancer 2010;9:383-7. [Crossref] [PubMed]
- Kim YB, Ham IH, Hur H, et al. Various ARID1A expression patterns and their clinical significance in gastric cancers. Hum Pathol 2016;49:61-70. [Crossref] [PubMed]
- Aso T, Uozaki H, Morita S, et al. Loss of ARID1A, ARID1B, and ARID2 Expression During Progression of Gastric Cancer. Anticancer Res 2015;35:6819-27. [PubMed]
- Abe H, Maeda D, Hino R, et al. ARID1A expression loss in gastric cancer: pathway-dependent roles with and without Epstein-Barr virus infection and microsatellite instability. Virchows Archiv 2012;461:367-77. [Crossref] [PubMed]
- Gong X, Yang L, Qin S. Progress in revealing the immunological mechanisms behind the anti-tumor activity of oxaliplatin. Chinese Clinical Oncology 2016;21:1031.
- Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut 2015;64:842-8. [Crossref] [PubMed]
- Lyu C, Zhang Y, Zhou X, et al. ARID1A gene silencing reduces the sensitivity of ovarian clear cell carcinoma to cisplatin. Exp Ther Med 2016;12:4067-71. [Crossref] [PubMed]
- Tong Q, Wang GB, Lu XM, et al. Expression of NF-κB, ICAM-1 and COX-2 in gastric carcinoma and their clinical significance. Chinese Journal of General Surgery 2005;14:7-739.
- Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 2011;43:1219-23. [Crossref] [PubMed]
- Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 2010;363:1532. [Crossref] [PubMed]
- Zeng Y, Liu Z, Yang J, et al. ARID1A is a tumor suppressor and inhibits glioma cell proliferation via the PI3K pathway. Head & Neck Oncology 2013;5:6.
- Inadomi K, Kusaba H, Matsushita Y, et al. Efficacy and Safety Analysis of Oxaliplatin-based Chemotherapy for Advanced Gastric Cancer. Anticancer Res 2017;37:2663-71. [Crossref] [PubMed]
- Luo B, Cheung HW, Subramanian A, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A 2008;105:20380-5. [Crossref] [PubMed]
- Han J, Meng Q, Xi Q, et al. PFKFB3 was overexpressed in gastric cancer patients and promoted the proliferation and migration of gastric cancer cells. Cancer Biomark 2017;18:249-56. [Crossref] [PubMed]
- Zhao B, Lin J, Rong L, et al. ARID1A promotes genomic stability through protecting telomere cohesion. Nat Commun 2019;10:4067. [Crossref] [PubMed]
- Williamson CT, Miller R, Pemberton HN, et al. ATR inhibitors as a synthetic lethal therapy for tumours deficient in ARID1A. Nat Commun 2016;7:13837. [Crossref] [PubMed]
- Alcindor T, Beauger N. Oxaliplatin: a review in the era of molecularly targeted therapy. Curr Oncol 2011;18:18-25. [Crossref] [PubMed]
- Wang L, Huang D, Jiang Z, et al. Akt3 is responsible for the survival and proliferation of embryonic stem cells. Biol Open 2017;6:850-61. [Crossref] [PubMed]
- Bitler BG, Fatkhutdinov N, Zhang R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin Ther Targets 2015;19:1419-22. [Crossref] [PubMed]
- Lee D, Yu EJ, Ham IH, et al. AKT inhibition is an effective treatment strategy in ARID1A-deficient gastric cancer cells. Onco Targets Ther 2017;10:4153-9. [Crossref] [PubMed]


