
A six-long noncoding RNA model predicts prognosis in lung adenocarcinoma
Introduction
Lung cancer is one of the most malignant tumors that poses great threat to the population health (1). According to surgery improving (2), molecular targeted drug developing and the application of immunotherapy, the overall survival (OS) of lung cancer has been significantly improved. Molecular targeted therapy has the characteristics of strong specificity and small side effects (3). The currently marketed molecular targeted drugs for non-small cell lung cancer (NSCLC) mainly include oncogene molecular targeted drugs, anti-angiogenesis drugs, immune targeted therapy drugs, and multi-target inhibitors. With the advancement of genetic testing technology and the wide application of small molecule tyrosine kinase inhibitors (TKIs), most patients with advanced NSCLC have achieved good therapeutic effects in the treatment of TKIs (4). However, some difficult issues still need to be resolved. Epidemiological studies show that lung adenocarcinoma (LUAD) has replaced lung squamous cell carcinoma (LUSC) as the main pathological type in lung cancers, but its pathogenesis and progressive mechanism remain unclear (5-7). Two-thirds of patients with LUAD are diagnosed at advanced stage, and they prone to earn poor prognosis for lacking effective individual therapy (8). Therefore, studying the molecular mechanism of LUAD to identify precise molecular typing markers is urgently needed.
It is well known that about 70% human genome would transcript into RNAs, of which protein-coding sequences account for less than 2%, the rest thousands of transcripts are non-coding RNAs (9,10). Long non-coding RNAs (lncRNAs) are defined more than 200 nucleotides, which occupied majority in non-coding RNAs (11). LncRNAs have been demonstrated involve in cell cycling controlling, cell differentiation mediating, epigenetic regulation and so on (12). Compared with protein-coding genes (PCGs), lncRNAs are composed of fewer exons to existing higher evolutional conservation (13). Moreover, lncRNAs show more stable feature against degradation as often forming secondary structure. These characters make lncRNAs easy to be detected in body fluids including blood and urine (14).
In recent years, the role of lncRNAs have also been widespread reported in tumorigenesis (15-18). LncRNA UCA1 is associated with poor prognosis in patients with gastric cancer (19). LINC00963 could promote tumorigenesis and radiation resistance of breast cancer by interacting with miR-324-3p (20). LncRNA PCNAP1 could enhance the replication of hepatitis B virus (HBV) and the occurrence of liver cancer (21). In ovarian cancer, lncRNA HOTTIP could indirectly up-regulate the expression of PD-L1, inhibit the activity of T cells and eventually accelerate the immune escape (22). And in lung cancer, the LCAT1-miR-4715-5p-RAC1/PAK1 axis plays an essential role during tumor progression and could be a potential therapeutic target (23). These findings suggest that the variation of lncRNAs are closely related to tumor prognosis. Up to now, most studies only focus on single lncRNA (24), there is still less work aim to analysis the correlation between lncRNAs and tumor prognosis systematically.
In this study, based on the Cancer Genome Atlas (TCGA) database, we analyzed the expression of all lncRNAs by Weighted Correlation Network analysis (WGCNA) to determine prognosis related module. Then, a six-lncRNA model with reliable prognostic value in LUAD was constructed by Cox proportional hazards regression analysis. In addition, we have conducted in-depth studies on the biological functions of this six-lncRNA model. Our results confirmed that this six-lncRNA model could be used to predict OS in LUAD independently, which could couple with traditional clinical prognostic factors to promote LUAD survival. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2436).
Methods
Patients and data pre-processing
A total of 479 LUAD samples containing clinical information were collected from TCGA database (https://portal.gdc.cancer.gov). Thirty-one patients with less than 30 days survival time were deleted, leaving 448 patients. We extracted 14,127 lncRNAs from the expression profile, and then screened 25% lncRNAs (n=3,532) with the largest variance differences for subsequent analysis. All data were filtered to reduce outliers. The flow chart of data collection and analysis were shown in Figure 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
WGCNA
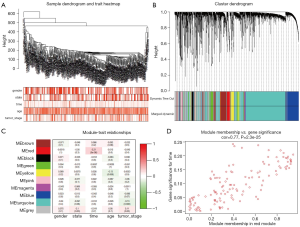
According to abline =650, 21 outlier samples were excluded, and 427 samples were remained. By choosing β =5 as the soft threshold, a WGCNA R package (25) was used to construct a weighted gene co-expression network. The topological overlap measurement (TOM) and the dynamic hybrid cutting method were used to identify co-expressed gene modules (26,27). Finally, removing the grey module, we got 9 modules, and the minimum number of lncRNAs in each clustering was set 30. According to the heatmap of module-trait relationships, we found the red module was significantly correlated with the prognosis. The 123 lncRNAs in the red module were used to construct a risk score model.
Construction of risk score model
The 427 samples were randomly divided into a training cohort (n=215) and a validation cohort (n=212) (Table S1). In order to verify the importance of lncRNAs in the red module, 123 lncRNAs in the training cohort were screened by univariate Cox proportional hazards regression, and 9 lncRNAs with P<0.05 were obtained. Then, lncRNAs with high correlation were removed by lasso regression. Finally, multivariate Cox proportional hazards regression was performed to determine the risk score model, the coefficients and hazard ratio (HR) values were obtained by Akaike information criterion (AIC). The calculation formula is as follows: risk score = Σ(C × EXPlncRNA). In our formula, EXPlncRNA represents the expression of six lncRNAs, and C represents the corresponding coefficient of multivariate Cox proportional hazards regression.
Validation and analysis of risk score model
Based on the median of risk score, the samples were divided into high-risk and low-risk groups. The receiver operating characteristic (ROC) curve was used to judge the prediction accuracy of risk score model. Kaplan-Meier curves were used to calculate OS and the statistical differences were determined by a log-rank test (28-30). Univariate and multivariate independent prognostic analysis were performed to determine whether the risk score could be distinguished from other clinical variables. Statistically, P<0.05 was set as significant differences.
Functional enrichment analysis
The entire LUAD samples were divided into two groups according to the median risk score. PCGs were screened by the cutoff criterion logFC >1.5, P<0.01. Finally, 689 differential genes were obtained. Functional enrichment analysis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of 689 differential genes were performed using the DAVID Bioinformatics Tool (https://david.ncifcrf.gov/, version 6.8) (31). Use the ClueGO package (http://apps.cytoscape.org/apps/cluego) in Cytoscape software to draw a network diagram of the biological process (BP). P<0.01 was used as the cutoff criterion for functional annotation of GO terms and KEGG pathways.
Gene set enrichment analysis (GSEA) was performed to explore potential pathways between high-risk and low-risk groups (32). Set the false discovery rate (FDR) <0.05 as the cutoff criterion.
Statistical analysis
All statistical tests were two-sided, and P<0.05 was considered statistically significant. Statistical analyses were conducted using R software (version 3.6.1, www.r-project.org).
Results
Data pre-processing of lncRNA profiles in LUAD
A total of 479 LUAD samples containing clinical information were downloaded from the TCGA database. According to the survival time, 31 patients were excluded, and 448 patients were remained. And the top 25% of lncRNAs (n=3,532) with the largest variance differences were screened for WGCNA analysis.
Identification of module related to the survival status of LUAD
To determine the expression characteristics of lncRNAs in LUAD, we constructed a co-expression network using WGCNA. After removing outliers, an adjacency matrix was constructed using 427 samples (Figure 2A). We choose β =5 as the soft power threshold to ensure that the correlation coefficient was close to 0.9 (Figure S1A,B). Then, 9 different-color co-expression modules were determined (Figure 2B). Finally, we found that this red module was significantly correlated with the survival status (cor =0.77, P<0.01) by analyzing the relationship between the modules and the clinical variables of LUAD (Figure 2C,D).
Identification of Cox proportional hazards regression model
The above 427 LUAD samples were randomly divided into two groups: training cohort (n=215) and validation cohort (n=212) (Table S1). In the training cohort, we performed univariate Cox and lasso regression analysis to obtained 9 lncRNAs based on 123 lncRNAs in the red module (Figure 3A). Finally, we constructed a risk score model through six lncRNAs by performing multivariate Cox analysis (Figure 3B, Table 1). Based on six lncRNAs, the risk score is calculated using the following formula: risk score = (−0.3057 × EXPVIM-AS1) + (0.9678 × EXPAC092811.1) + (1.0829 × EXPNFIA-AS1) + (−0.3505 × EXPAL035701.1) + (3.9378 × EXPAC079336.4) + (−0.2810 × EXPAL121790.2). At the same time, based on the overall data of TCGA-LUAD, we plotted the Kaplan-Meier curve of six lncRNAs (Figure 3C,D,E,F,G,H).
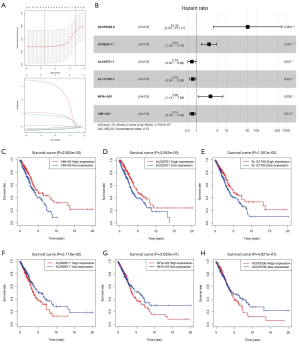
Table 1
| Symbol | Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|---|
| HR | 95% CI of HR | P value | HR | 95% CI of HR | P value | Coefficient | ||
| VIM-AS1 | 0.714 | 0.573–0.889 | 0.003 | 0.737 | 0.580–0.936 | 0.012 | −0.30569451 | |
| AC092811.1 | 2.559 | 1.388–4.719 | 0.003 | 2.632 | 1.450–4.777 | 0.001 | 0.96776812 | |
| NFIA-AS1 | 3.488 | 1.468–8.287 | 0.005 | 2.953 | 1.136–7.680 | 0.026 | 1.08294756 | |
| AL035701.1 | 0.663 | 0.480–0.916 | 0.013 | 0.704 | 0.516–0.962 | 0.027 | −0.35051832 | |
| AC079336.4 | 28.828 | 1.397–594.845 | 0.030 | 51.304 | 3.698–711.708 | 0.003 | 3.93777712 | |
| AL121790.2 | 0.778 | 0.612–0.988 | 0.039 | 0.755 | 0.594–0.960 | 0.022 | −0.28098658 | |
LncRNAs, long non-coding RNAs; HR, hazard ratio; CI, confidence interval.
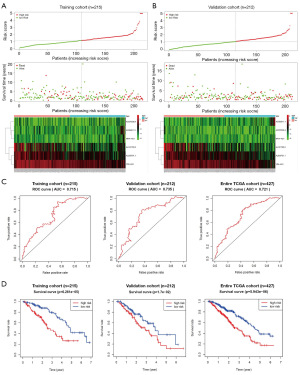
Prognostic efficiency and validation of six-lncRNA model
Based on the median risk score, we divided the training cohort into high-risk and low-risk groups. It could be seen from the scatter plot of Figure 4A, the mortality of high-risk patients was significantly higher than low-risk patients. From the heatmap of Figure 4A, as the risk score increases, three lncRNAs expression (VIM-AS1, AL035701.1 and AL121790.2) were gradually decreased, and three lncRNAs expression (AC092811.1, NFIA-AS1 and AC079336.4) were gradually increased. Using the same classification method, we divided the validation cohort into high-risk and low-risk groups. We also found a significant increase of mortality in high-risk patients (Figure 4B). In addition, our results indicated that the 5-year survival areas under the ROC curve were 0.715, 0.735 and 0.721 in the training cohort, validation cohort and the entire TCGA data, which imply that this six-lncRNA model has good predictive value (Figure 4C). Finally, we found that the 5-year survival rates were significantly worse in the high-risk group compared with the low-risk group (all P<0.001) (Figure 4D).
Independent prognostic ability and prognostic value of six-lncRNA model
Based on the training cohort, univariate and multivariate independent prognostic analysis were used to analyze the correlation of gender, age, TNM stage, T stage, N stage, M stage, risk score and prognosis. Through univariate independent prognostic analysis, we found that risk score was a powerful variable related to prognosis (P<0.001) (Figure 5A, Table 2). By adding other clinical variables for multivariate independent prognostic analysis, we found that risk score could also be used as an independent prognostic variable (P<0.001) (Figure 5B, Table 2).
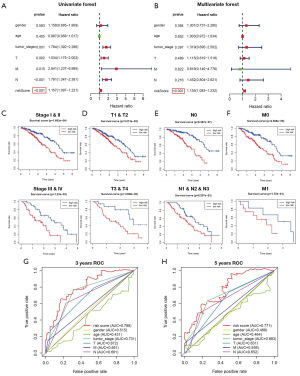
Table 2
| Variables (n=215) | Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|---|
| HR | 95% CI of HR | P value | HR | 95% CI of HR | P value | ||
| Gender | 1.158 | 0.685–1.959 | 0.583 | 1.301 | 0.737–2.295 | 0.364 | |
| Age | 0.987 | 0.958–1.017 | 0.405 | 1.003 | 0.972–1.034 | 0.852 | |
| Tumor stage | 1.784 | 1.392–2.286 | 4.70E−06 | 1.319 | 0.695–2.502 | 0.397 | |
| T | 1.534 | 1.175–2.002 | 0.002 | 1.115 | 0.819–1.518 | 0.489 | |
| M | 2.941 | 1.237–6.995 | 0.01466 | 0.816 | 0.140–4.776 | 0.822 | |
| N | 1.791 | 1.347–2.381 | 6.05E−05 | 1.452 | 0.804–2.621 | 0.216 | |
| Risk score | 1.157 | 1.097–1.221 | 9.53E−08 | 1.155 | 1.083–1.232 | 1.14E−05 | |
HR, hazard ratio; CI, confidence interval.
Throughout the TCGA database, we verified the independent prognostic capabilities of risk score. According to TNM staging, patients were divided into early stage (stage I & II) and advanced stage (stage III & IV) for analysis. We found that the risk score can successfully predict the survival outcome in two subgroups (all P<0.01) (Figure 5C). According to the T stage, the patients were divided into the highly differentiated group (T1 & T2) and the poorly differentiated group (T3 & T4) for analysis. We found that both groups were significantly different (P<0.01, P=0.01) (Figure 5D). According to the analysis of lymph node metastasis (N0, N1 & N2 & N3) and distant metastasis (M0, M1), we found that in the N0, N1 & N2 & N3 or M0 stage, the risk score could successfully predict the survival outcome (all P<0.01) (Figure 5E,F). However, we have not found significant difference in the M1 stage, which may be related with the few samples. But we found that the high-risk group in the M1 stage had a lower survival rate than the low-risk group (Figure 5F). Similarly, subgroup analysis in age (<66, ≥66) and gender (male, female) showed that the risk score could predict the survival outcome (Figure S2A,B). These results indicated that the predictive ability of six-lncRNA model was not affected by gender, age, TNM stage, T stage, N stage, and M stage. In addition, based on the training cohort, we found that the 3-year and 5-year areas of six-lncRNA model under the ROC curve were higher (AUC =0.798, AUC =0.771) than the other clinical variables (Figure 5G,H).
Functional enrichment analysis
The biological function of lncRNAs is still unknown. Therefore, in order to accurate evaluated the biological function of this six-lncRNA model, we analyzed the function of 689 differential genes according to the high-risk and low-risk groups of risk score (Figure S3A). As shown in Figure 6A, BPs were mainly involved in the cell cycle process and DNA metabolic process. The cellular components (CCs) were mainly enriched in nuclear chromosome part and chromatin (Figure 6B). Enriched molecular functions (MFs) were mainly enriched in chromatin binding and cell adhesion molecule binding (Figure 6C). KEGG functional analysis found that the main enrichment was spliceosome, cell cycle and DNA replication (Figure 6D).
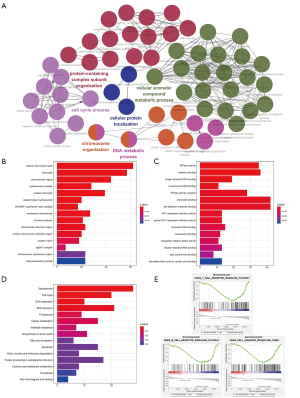
The functional GSEA showed that the high-risk group were highly enriched in proteasome and protein export (Figure S3B), and the low-risk group were highly enriched in cell adhesion molecules and T/B cell receptor signaling pathway (Figure 6E).
Discussion
In the past, most researches concentrated on studying the role of PCGs in cellular behavior regulation (33-35). However, some cellular behaviors alterations have been found not mediated by PCGs, either linked to “gene deserts” regions. It is reported that only 2% of the human genome encodes proteins, which further supports the potential regulation role of “gene deserts” regions in cellular behaviors (36,37). As products of “gene deserts” regions, lncRNA dysfunctions have been presented in various tumors and are closely related with progressing (38,39). But there is still rare work try to analysis the correlation between lncRNAs and tumor prognosis systematically (24). In this work, we employed the LUAD database from TCGA to construct a six-lncRNA prognostic model and investigated its prognostic evaluation efficiency by the following steps: a prognosis module was screened through WGCNA, and then a six-lncRNA model was identified by constructing multivariate Cox proportional hazards regression and verified by validation cohort. Finally, we have conducted in-depth studies on the biological functions of this six-lncRNA model.
This six-lncRNA model is consisted by VIM-AS1, AL035701.1, AL121790.2, AC092811.1, NFIA-AS1 and AC079336.4. Our study found that among these lncRNAs, the low expression of 3 lncRNAs (VIM-AS1, AL035701.1 and AL121790.2) and the high expression of 3 lncRNAs (AC092811.1, NFIA-AS1 and AC079336.4) are related with the poor prognosis in LUAD (Figure 3C,D,E,F,G,H). It is reported that VIM-AS1 could promote the progression and metastasis of colorectal cancer by inducing EMT (40). In addition, the abnormal expression of VIM-AS1 in cumulus cells during embryonic development is crucial for oocyte growth (41). Although the other five lncRNAs have not been reported in LUAD-related research, more works are needed to verify this finding in the future. In addition, based on the TNM stage, T stage, N stage and M stage, we have verified that the independent prognostic capabilities of six-lncRNA model is statistically significant (all P<0.001, Figure 5C,D,E,F). And we also found that the 5-year areas of the six-lncRNA model (AUC =0,771) under the ROC curve was higher than other clinical variables, such as gender (AUC =0.488), age (AUC =0.464), tumor stage (AUC =0.693), T stage (AUC =0.631), N stage (AUC =0.652) and M stage (AUC =0.559) (Figure 5H). These results indicate that the six-lncRNA model shows a higher-risk detection efficiency as contrasted to other clinical variables.
To determine the biological function of the six-lncRNA model, we analyzed the function of 689 differential genes obtained by high-risk and low-risk groups. We found that these differential genes were almost related with cell cycle process, cell adhesion molecule binding and the cell cycle and DNA replication pathway. Among them, cell adhesion regulation is a key factor for tumor invasion and occurrence (42), cell cycle processing and DNA replication also been shown related to tumor development and occurrence (29,30). GSEA analysis found that the low-risk group was mainly enriched in cell adhesion molecules and T/B cell receptor signaling pathway, which imply that the immune system is involve in suppressing the malignant processes of tumor.
Conclusions
Epidemiological statistics show that the incidence of various types of lung cancer has changed significantly compared with the past. LUAD has replaced LUSC as the main type of lung cancers (7). It shown that even LUAD patients exhibit same clinical stage and nearly pathological subtype, they may bear different prognosis and recurrence risk after surgery. In this study, we constructed a Cox proportional hazards regression model consisting of six-lncRNA, which could help physicians to précising subtype LUAD patients and give them more individual treatment.
Acknowledgments
The authors are grateful to all study participants.
Funding: The research was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2436
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2436
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2436). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yoda S, Dagogo-Jack I, Hata AN. Targeting oncogenic drivers in lung cancer: Recent progress, current challenges and future opportunities. Pharmacol Ther 2019;193:20-30. [Crossref] [PubMed]
- Petrella F, Spaggiari L. Therapeutic options following pneumonectomy in non-small cell lung cancer. Expert Rev Respir Med 2016;10:919-25. [Crossref] [PubMed]
- Reungwetwattana T, Dy GK. Targeted therapies in development for non-small cell lung cancer. J Carcinog 2013;12:22. [Crossref] [PubMed]
- Sa H, Ma K, Gao Y, et al. Predictive Value of Tumor Mutation Burden in Immunotherapy for Lung Cancer. Zhongguo Fei Ai Za Zhi 2019;22:380-4. [PubMed]
- Inamura K. Clinicopathological Characteristics and Mutations Driving Development of Early Lung Adenocarcinoma: Tumor Initiation and Progression. Int J Mol Sci 2018;19:1259. [Crossref] [PubMed]
- Matěj R, Rohan Z, Němejcová K, et al. Molecular pathology of lung cancer in routine diagnostic practice: 2017 update. Cesk Patol 2017;53:159-66. [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Peng W, Wang J, Shan B, et al. Diagnostic and Prognostic Potential of Circulating Long Non-Coding RNAs in Non Small Cell Lung Cancer. Cell Physiol Biochem 2018;49:816-27. [Crossref] [PubMed]
- Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012;22:1775-89. [Crossref] [PubMed]
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature 2012;489:101-8. [Crossref] [PubMed]
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013;152:1298-307. [Crossref] [PubMed]
- Chen JH, Zhou LY, Xu S, et al. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int 2017;17:64. [Crossref] [PubMed]
- Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915-27. [Crossref] [PubMed]
- Su M, Xiao Y, Tang J, et al. Role of lncRNA and EZH2 Interaction/Regulatory Network in Lung Cancer. J Cancer 2018;9:4156-65. [Crossref] [PubMed]
- Yao G, Chen K, Qin Y, et al. Long Non-coding RNA JHDM1D-AS1 Interacts with DHX15 Protein to Enhance Non-Small-Cell Lung Cancer Growth and Metastasis. Mol Ther Nucleic Acids 2019;18:831-40. [Crossref] [PubMed]
- Wu W, Zhao Y, Gao E, et al. LncRNA DLEU2 accelerates the tumorigenesis and invasion of non-small cell lung cancer by sponging miR-30a-5p. J Cell Mol Med 2020;24:441-50. [Crossref] [PubMed]
- Huang N, Guo W, Ren K, et al. LncRNA AFAP1-AS1 Supresses miR-139-5p and Promotes Cell Proliferation and Chemotherapy Resistance of Non-small Cell Lung Cancer by Competitively Upregulating RRM2. Front Oncol 2019;9:1103. [Crossref] [PubMed]
- Zhao W, Zhang LN, Wang XL, et al. Long noncoding RNA NSCLCAT1 increases non-small cell lung cancer cell invasion and migration through the Hippo signaling pathway by interacting with CDH1. FASEB J 2019;33:1151-66. [Crossref] [PubMed]
- He X, Wang J, Chen J, et al. lncRNA UCA1 Predicts a Poor Prognosis and Regulates Cell Proliferation and Migration by Repressing p21 and SPRY1 Expression in GC. Mol Ther Nucleic Acids 2019;18:605-16. [Crossref] [PubMed]
- Zhang N, Zeng X, Sun C, et al. LncRNA LINC00963 Promotes Tumorigenesis and Radioresistance in Breast Cancer by Sponging miR-324-3p and Inducing ACK1 Expression. Mol Ther Nucleic Acids 2019;18:871-81. [Crossref] [PubMed]
- Feng J, Yang G, Liu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics 2019;9:5227-45. [Crossref] [PubMed]
- Shang A, Wang W, Gu C, et al. Long non-coding RNA HOTTIP enhances IL-6 expression to potentiate immune escape of ovarian cancer cells by upregulating the expression of PD-L1 in neutrophils. J Exp Clin Cancer Res 2019;38:411. [Crossref] [PubMed]
- Yang J, Qiu Q, Qian X, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer 2019;18:171. [Crossref] [PubMed]
- Lin T, Fu Y, Zhang X, et al. A seven-long noncoding RNA signature predicts overall survival for patients with early stage non-small cell lung cancer. Aging (Albany NY) 2018;10:2356-66. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Li A, Horvath S. Network module detection: Affinity search technique with the multi-node topological overlap measure. BMC Res Notes 2009;2:142. [Crossref] [PubMed]
- Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 2008;24:719-20. [Crossref] [PubMed]
- Ramos M, Schiffer L, Re A, et al. Software for the Integration of Multiomics Experiments in Bioconductor. Cancer Res 2017;77:e39-42. [Crossref] [PubMed]
- Singhal S, Vachani A, Antin-Ozerkis D, et al. Prognostic implications of cell cycle, apoptosis, and angiogenesis biomarkers in non-small cell lung cancer: a review. Clin Cancer Res 2005;11:3974-86. [Crossref] [PubMed]
- Ho ST, Lin CC, Tung YT, et al. Molecular Mechanisms Underlying Yatein-Induced Cell-Cycle Arrest and Microtubule Destabilization in Human Lung Adenocarcinoma Cells. Cancers (Basel) 2019;11:1384. [Crossref] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44-57. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Seidl C, Panzitt K, Bertsch A, et al. MicroRNA-182-5p regulates hedgehog signaling pathway and chemosensitivity of cisplatin-resistant lung adenocarcinoma cells via targeting GLI2. Cancer Lett 2020;469:266-76. [Crossref] [PubMed]
- Bai Y, Xiong L, Zhu M, et al. Co-expression network analysis identified KIF2C in association with progression and prognosis in lung adenocarcinoma. Cancer Biomark 2019;24:371-82. [Crossref] [PubMed]
- Zeng F, Wang Q, Wang S, et al. Linc00173 promotes chemoresistance and progression of small cell lung cancer by sponging miR-218 to regulate Etk expression. Oncogene 2020;39:293-307. [Crossref] [PubMed]
- Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010;463:899-905. [Crossref] [PubMed]
- Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134-40. [Crossref] [PubMed]
- Peng F, Wang R, Zhang Y, et al. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer 2017;16:98. [Crossref] [PubMed]
- Xie Y, Zhang Y, Du L, et al. Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non-small cell lung cancer. Mol Oncol 2018;12:648-58. [Crossref] [PubMed]
- Rezanejad Bardaji H, Asadi MH, Yaghoobi MM. Long noncoding RNA VIM-AS1 promotes colorectal cancer progression and metastasis by inducing EMT. Eur J Cell Biol 2018;97:279-88. [Crossref] [PubMed]
- Bouckenheimer J, Fauque P, Lecellier CH, et al. Differential long non-coding RNA expression profiles in human oocytes and cumulus cells. Sci Rep 2018;8:2202. [Crossref] [PubMed]
- Olajuyin AM, Olajuyin AK, Wang Z, et al. CD146 T cells in lung cancer: its function, detection, and clinical implications as a biomarker and therapeutic target. Cancer Cell Int 2019;19:247. [Crossref] [PubMed]



