
Identification of CDK1 as a candidate marker in cutaneous squamous cell carcinoma by integrated bioinformatics analysis
Introduction
Cancer is a major threat to human health and a global economic burden. The economic impact associated with anti-cancer therapies can largely affect treatment access and continuity, increasing the likelihood of poor prognoses (1). Cutaneous squamous cell carcinoma (cSCC) is a relatively common cancer that accounts for nearly 50% of non-melanoma skin cancer cases (2). In addition to environmental exposure, a variety factors such as aging, human papilloma virus infection, immunosuppression, genetic factors and in particular, ultraviolet radiation are all associated with increased cSCC risk. At present the preferred treatment of cSCC is radical resection. Although most cSCC patients who undergo cruative surgery exhibit favorable prognoses, a minority develop metastasis or recurrence that is potentially fatal. For patients with advanced cSCC, due to the lack of large-scale clinical trials, clinicians have to rely on the efficacy evidence of other tumor types (such as head and neck mucosal squamous cell carcinoma) to guide treatment plan. Systemic therapy, like platinum chemotherapy, were used in advanced cSCC patients population (3,4). Genetic alterations have shown to play a key role in the resistance to anticancer drugs in oncology patients, demonstrating an urgent need to identify key genes that can serve as therapeutic targets and biomarkers for monitoring the response to treatment (5). Recently, EGFR inhibitors, cemiplimab and pembrolizumab which aim at biomarkers were used for patients with advanced cSCC.
The development of microarray and high-throughput sequencing technologies has facilitated the identification of biomarkers related to tumor occurrence and progression (6-8). Unlike head and neck squamous cell carcinoma, relatively few marker genes have been identified in cSCC (9,10).
In recent research, Wei et al. (11) suggested that EGR3 had the potential to help in the diagnosis and predictive treatments of CSCC by bioinformatics, but the samples in the study are not from the same organization or the same GEO profiles. To address this deficiency, bioinformatic analysis was performed in our study for providing more reliable research evidence for the early diagnosis and prevention of cSCC the development and progression. In our study, we used the same tissue-derived specimens, and compare the different genes in the same GEO profile which helped to avoid partial bias. In addition, we validated the results of bioinformatics in order to enhance the reliability of the study.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2945).
Methods
Microarray analysis and identification of DEGs
The three microarray datasets of primary cSCC and normal tissues (GSE117247, GSE32979, and GSE98767) were obtained from the NCBI GEO database. The GSE117247 data was based on the Affymetrix Human Genome U95 v2 array (GPL8300 platform), derived from eight primary cSCC and nine normal skin samples. The GSE32979 microarray data were obtained using the Illumina Human-6 v2.0 expression bead chip (GPL6102 platform) from 15 cSCC and 13 normal skin samples. GSE98767 data were obtained using the Illumina Human HT-12 v4.0 expression bead chip (GPL10558 platform) from nine primary cSCC and nine normal tissue samples. The integration and analysis of all three datasets was performed with GEO2R, a web-based analysis tool [https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE98767], and saved.. Genes differentially expressed between cSCC and normal tissues were screened according to a standard P value of <0.05, and fold change (FC) in gene expression >0 (|logFC| >0). The DEGs were categorised into Venn diagrams using a web-based application (http://bioinformatics.psb.ugent.be/webtools/Venn/). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
GO and KEGG pathway enrichment analyses
The online gene function annotation tool, Database for Annotation, Visualization, and Integrated Discovery (DAVID), (https://david.ncifcrf.gov/), was used to analyze the functions and pathway enrichment of candidate DEGs. The cut-off for calculating the degree of enrichment was P<0.05.
PPI networks, clustering, and identification of hub genes
Proteins corresponding to DEGs and a PPI network diagram were obtained with STRING (https://string-db.org), an online tool for identifying functional protein association networks. Candidate genes were evaluated with the Cytoscape (Version 3.7.2) software to analyze interactions between genes. All analyzed genes and modules of PPI network underwent further screening with the MCODE plug-in [degree cut off =2 (12), node score cut off =0.2, k-core =2, max. Depth =100].
Statistical analysis
Data were analyzed with the paired-samples t test using Prism 6 software (GraphPad, La Jolla, CA, USA). P<0.05 was considered statistically significant.
Immunohistochemical (IHC) analysis
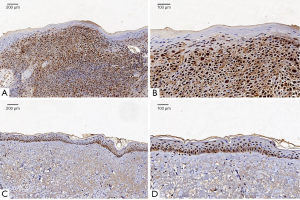
All samples (including normal tissues adjacent to carcinoma and cSCC tissues) obtained from the 20 cSCC patients were fixed in 10% formalin for 24 h at room temperature and embedded in paraffin. The paraffin-embedded tissues were serially cut into 5µm sections and heated in an oven at 60 °C for 2 h. The tissue sections were dewaxed, antigen-repaired according to high temperature and high pressure. After antigen repair, the sections were rinsed with PBS for 3 times, and incubated with Cdc2 p34 (200 µg/mL; 1:200; cat. no. sc-54; Santa Cruz), BUB1(B-3) (200 µg/mL; 1:200; cat. no. sc-365685; Santa Cruz), MAD2(C-10) (200 µg/mL; 1:200; cat. no. sc-374131; Santa Cruz) for 18 h at 4 °C, respectively. The sections in incubation box was rinsed with PBS 3 times for 5 minutes. The excess PBS was removed, and the m-IgGκ BP-HRP (cat. no. sc-516102, Santa Cruz) was added before incubation at 37 °C for 30 min. And then the sections were rinsed with PBS for 3 times. DAB Color-substrate solution (cat. no. K5007; DAKO) was added for 3–5 min, followed by rinsing with running water for 5–10 min to stop the color development. Then, the tissue sections were counterstained with hematoxylin for 1 min and differentiated with 0.5% hydrochloric alcohol for 2 seconds. The sections were dehydrated and mounted for microscopic examination at room temperature. The signal was visualized with Light microscopy (Olympus BX43; Olympus Corporation) (magnification, ×200) was performed, and the presence of brown granules in the cytoplasm was considered a positive signal.
Results
Identification of DEGs in cSCC
Gene expression profiles of 32 cSCC and 31 normal skin tissue samples were compared in this study. GEO2R analysis yielded 2749, 8062, and 8472 identified DEGs in GSE117247, GSE32979, GSE98767 datasets, respectively. Among the 756 genes co-expressed across the three datasets, 219 were shown to be upregulated (logFC >0) and 116 were downregulated (logFC <0) in cSCC compared to normal tissue (Figure 1 and Table 1).
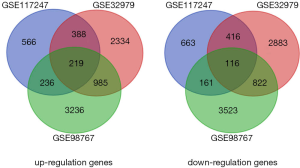
Table 1
| DEGs | Gene names |
|---|---|
| Down-regulated genes | ABR, MAOA, GNA11, ARAP1, KRT15, ITSN1, DCT, KCTD2, CRTAP, ABCA6, LGALS3, LPCAT4, KIAA0895, MAN2B2, MBP, CDH19, CTIF, ESRRG, CNGA1, CHKB, MAPRE3, GCHFR, ATG9A, CAMK2G, LSS, ID4, IQGAP1, GSTM3, ACVR1B, AMT, AHNAK2, CDK10, ITPR3, GHR, DNM1, ALDH6A1, DIXDC1, ARHGAP12, MXRA5, DAG1, MIA, CCS, CTNNBIP1, HMGN2, ASMTL, MZF1, ABCA2, FXYD1, FASTK, EFS, ASPA, AGTR1, EPHX2, BPTF, DST, ABCA3, N4BP2L2, BCL2, ZBTB20, FAM193A, MAN2C1, EEF1D, CRELD1, GPC3, AKAP8, ABCA5, MACF1, CAPN3, DIAPH2, CRYAB, GSN, HOXA10, SLIT3, ITPR2, GOSR1, EZH1, CIC, HTT, FAM172A, MON2, ARMCX6, ARHGEF10, ATG14, B3GAT3, AHDC1, MITF, GABBR1, FYN, BARD1, CBX7, CYB5R3, ICA1, LAMB2, NBAS, FABP7, ARHGEF7, MAN2A2, MAU2, DDX42, FBLN1, BMP4, ESD, LAMA5, TSPOAP1, ECM2, LYRM9, ACTR1B, SLF2, BAP1, FAM189A2, CADM1, DNAJB12, HOXC6, DKFZP586I1420, ERCC2 |
| Up-regulated genes | ATP5F1, LMAN1, LMNB2, FAM98A, FOXM1, CDK1, GCLM, ABCF2, GSTP1, KIF14, MAD2L1, EIF4A3, GOT2, MANF, EIF4E2, C1QBP, CASP1, GCH1, MELK, GPX2, NDC80, AP2S1, MYC, CCNA2, ISCA1, ALAS1, CRLF3, AMPD3, CKS2, BCR, CCNB2, MPHOSPH6, HIST1H2AC, DDX21, AZIN1, CBX3, NCK1, PUDP, CSTB, GTF2E2, CCNF, HDAC1, CNIH1, BLZF1, EIF3I, CASP4, DYNLT3, CSNK2A1, TYMP, ABLIM1, ACTR2, ATOX1, BID, COX5A, ATP6V0D1, GNAI3, FCGR2B, ABCG1, LYPLA1, NAPG, HMOX2, ATP6V0B, AKIRIN2, KIF11, CCT2, ARF4, MED8, DSC2, MID1, DBF4, AHCY, MAPK14, ARPP19, MPZL1, NOP16, CAP1, CDC42, EREG, CDC20, GART, DNAJB6, H2AFZ, CCNT2, BUB1, DFNA5, HNRNPC, DUSP6, MRPL3, MAPK6, EIF5, TPGS2, DPH2, FEN1, HPRT1, HIF1A, ACTB, CACYBP, CSRP2, CUX1, AIM2, HDGF, ATP1B3, E2F3, GARS, H2AFY, HSD17B2, CSTF1, BAK1, PUM3, KPNA2, MRPS12, ACOT7, KIF2A, GLA, ARPC3, APOBEC3B, AURKA, AP3B1, C6orf62, FAM189B, IFI16, CEBPD, DDX39A, ERCC1, DUSP14, EIF2B2, ARFGEF2, UBE2K, ARPC1A, HIST1H2BE, FGFBP1, CDH3, EI24, COPB2, GGH, HMOX1, CTNS, GADD45A, GTF2A2, COPE, FSCN1, CFL1, GBP1, ATP5J, ACVR1, EIF4EBP1, GSPT1, GGCT, HIST1H2BK, DLGAP5, ENO1, GOSR2, ADAM17, EIF4H, CTSA, APEX2, DNM1L, CDC42EP1, KIAA0101, BCL10, CDA, EPHB2, CKAP4, AGFG1, HMGB3, HOMER3, GK, ASCC3, EPHA2, CYFIP1, DYNLT1, GNG5, MTHFD2, CSK, MORF4L2, BZW1, KYNU, ETV4, CCL20, DNAJA1, KIF2C, CYB561D2, ADAM10, GNA15, CLCA2, AK2, ILF3, CSNK1A1, CCNC, DCTN6, BDH1, CD24, CSTF2, DNMT1, CAPRIN1, HOMER1, HK2, NCBP1, ADK, CA2, GRN, FGFR1OP, FTSJ1, FLAD1, DSG3, BUB1B, FMO2, ALG3, EXOG, ATP12A, GIPC1, ATP6V1D, EIF4G2, ATP2A2, CASK, ATP6V0E1, AMD1, CTSC |
cSCC, Cutaneous Squamous Cell Carcinoma; DEGs, Differentially Expressed Genes.
Analysis of GO terms for DEGs in cSCC
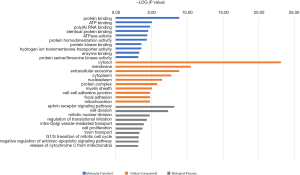
GO analysis of the DEGs was undertaken using P<0.05 as the cut off. Under the biological process (BP) domain, DEGs were mainly enriched in the Ephrin receptor signaling pathway and cell division. The top three processes under Cellular Components (CC) domain were cytosol, membrane, and extracellular exosome. Protein-, ATP, poly(A) RNA-, and identical protein binding were the highest ranked processes under the Molecular function (MF) domain (Figure 2).
KEGG pathway enrichment analysis
Signaling pathways associated with the DEGs were examined, with P<0.05 as the cut off. Identified candidate pathways, linked to DEGs were shown to be involved in various BPs, including infection (epithelial cell signaling in Helicobacter pylori infection, pathogenic escherichia coli infection, hepatitis C), cell proliferation and death (cell cycle, oocyte meiosis), immunity (Gamma R-mediated phagocytosis), and metabolism (lipolysis regulation in adipocytes) (Table 2). The data collectively suggests that the main pathways involved in the development of cSCC are those associated with metabolism.
Table 2
| Pathway | Gene count | P value | Genes |
|---|---|---|---|
| Cell cycle | 12 | 3.29E-03 | MAD2L1, BUB1, BUB1B, CDC20, CDK1, E2F3, CCNB2, HDAC1, DBF4, CCNA2, GADD45A, MYC |
| Epithelial cell signaling in Helicobacter pylori infection | 9 | 1.97E-03 | CDC42, ATP6V0E1, ADAM10, MAPK14, ADAM17, ATP6V0D1, CSK, ATP6V1D, ATP6V0B |
| Oocyte meiosis | 9 | 3.29E-02 | CDK1, MAD2L1, CCNB2, CAMK2G, BUB1, CDC20, AURKA, ITPR3, ITPR2 |
| Fc gamma R-mediated phagocytosis | 8 | 4.29E-02 | ARPC1A, CDC42, DNM1L, ARPC3, FCGR2B, GSN, CFL1, DNM1 |
| Pathogenic Escherichia coli infection | 6 | 4.30E-02 | ARPC1A, ACTB, CDC42, ARPC3, FYN, NCK1 |
cSCC, cutaneous squamous cell carcinoma.
PPI network and modular analyses
An integrated analysis of the 335 candidate genes was performed using the STRING database, generating a PPI network diagram which excluded 332 nodes and 1,307 edges. Pathway analyses using GO and KEGG (Figure 3) revealed that the DEGs were mainly enriched in pathways related to Epithelial cell signaling in H. pylori infection and the cell cycle. Analysis of the DEGs with Cytoscape software and the MCODE plug-in was performed. The top two functional modules are shown in Figure 4. Module 1 exhibited 20 nodes and 185 edges, whilst Module 2 showed 55 nodes and 391 edges. GO and KEGG pathway enrichment analyses of the DEGs in both modules displayed 20 upregulated and no downregulated genes in Module 1 (Figure 4A). However, Module 2 (Figure 4B), contained 47 upregulated genes, and 8 that were downregulated. The genes in both modules were mainly related to cell cycle functions (Table 3).
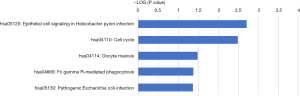
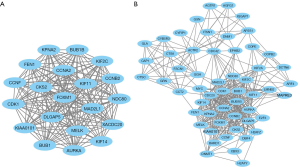
Table 3
| Term | Description | Count | P value |
|---|---|---|---|
| Module 1 | |||
| GO:0051301 (Biological process) | Cell division | 14 | 2.7×1018 |
| GO:0000776 (Cellular component) | Kinetochore | 5 | 1.3×106 |
| GO:0019901 (Molecular function) | Protein kinase binding | 6 | 4.8×105 |
| hsa:04110 (KEGG pathway) | Cell cycle | 7 | 6.0×109 |
| Module 2 | |||
| GO:0051301 (Biological process) | Cell division | 17 | 6.7×1015 |
| GO:0005829 (Cellular component) | Cytosol | 29 | 2.2×108 |
| GO:0005515 (Molecular function) | Protein binding | 46 | 5.0×107 |
| hsa:04110 (KEGG pathway) | Cell cycle | 10 | 2.2×109 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
We also analyzed the top 30 hub genes with the Betweennes, MCC, MNC, Degree, EPC, BottleNeck, EcCentricity, Closenes, Radiality, and stress methods. The 17 hub genes identified by these nine statistical methods were KIF2C, CDC42, AURKA, MAD2L1, MYC, CDK1, FEN1, H2AFZ, BUB1, BUB1B, CKS2, CDC20, CCT2, ACTR2, ACTB, MAPK14, and HDAC1. These DEGs are critically involved in the pathogenesis and development of cSCC.
Expression of CDK1 in cSCC
Based on the above results, CDK1 expression in cSCC was analyzed in tumor and normal tissues via IHC analysis. The results are shown in Figure 5A,B, where cSCC tissue is indicated by the presence of positively stained brown nuclei. CDK1 expression was higher in cSCC compared to normal skin tissue (Figure 5C,D), consistent with results obtained by integrating the GEO profiles. These results suggested that CDK1 may serve as a new biomarker for the diagnosis and prognosis of cSCC. The expression of MAD2L1 and BUB1 protein in cSCC showed no significant difference compare with that in normal tissues.
Discussion
In this study, we used integrated bioinformatics analysis to screen 756 genes in three GEO expression profiles, including 219 that were shown to be upregulated and 116 that were downregulated in cSCC, compared to normal tissue. In the BP domain of the GO analysis, most DEGs were enriched in Ephrin receptor signaling and cell division, which have been implicated in angiogenesis, stem cell differentiation, and cancer. Cytosol and protein binding were enriched in the CC and MF domains, respectively. KEGG analysis also revealed Epithelial cell signaling in H. pylori infection, cell cycle, and oocyte meiosis as the highest ranking pathways associated with the DEGs. In the PPI network, two co-expression clusters analyzed with Cytoscape MCODE were found to comprise 20 and 55 genes, respectively. Cell division was the most significant BP in Modules 1 and 2.
Most cancers are associated with genetic mutations involved in cell cycle regulation or aberrant activities of cyclin-dependent kinases (CDK), cyclins, and CDK inhibitors (8-10), which are normally required in regulating cell growth. CDK1 encodes a Ser/Thr kinase that plays an essential part in the eukaryotic cell cycle and was widely found to be overexpressed in oral SCC (13), liver cell carcinoma (14), epithelial ovarian cancer (15), and breast cancer (15). In contrast, gene silencing of CDK1 has exhibited therapeutic potential in epithelial ovarian cancer (16), breast cancer (17), and malignant pleural mesothelioma (18).
Mitotic arrest defect 2 ligand 1 (MAD2L1) encodes a cell cycle checkpoint protein that modulates anaphase-promoting complex/cyclosome (APC/C) to ensure the correct arrangement of chromosomes on the metaphase plate during cell division (19). MAD2L1 expression has recently been linked to tumor stage and prognosis (20). Additionally, high expression of MAD2L1 has been observed in breast cancer (21), gastric cancer (22), and other malignancies.
Mutation of Cell Division Cycle protein (CDC)20 leads to aberrant mitotic arrest and chromosome segregation (23). The activation of APC/C-CDC20 enhances the sensitivity of cancer cells to radiotherapy and chemotherapy (24,25). CDC20 overexpression was recently reported in gastric cancer, colorectal cancer, hepatocellular carcinoma, cervical cancer, and bladder cancer. Moreover, CD20 overexpression was highly correlated with poor prognoses (26-29). Conversely, downregulation of CDC20 slowed growth and colony formation in small and non-small cell lung carcinoma cells (30).
The budding uninhibited by benzimidazoles 1 homolog beta (BUB1B) gene encodes a kinase that is associated with cell cycle checkpoints and chromosome segregation (31). BUB1B binds to CDC20 during the G2 phase of mitosis to inhibit APC/C activity and cell cycle progression (32). The upregulated BUB1B level was found to be highly correlated with unfavorable prognosis in glioblastoma (33) and hepatocellular carcinoma (34). Furthermore, its overexpression is thought to enhance the resistance of glioblastoma to radiotherapy (35).
To date, few studies have undertaken detailed investigations into the precise role of CDK1 in the development and progression of cSCC. Our findings demonstrate that CDK1 was highly expressed in cSCC, providing a sound rationale for future studies in rapidly establishing the role and mechanism of CDK1 in cSCC. The development of cSCC is complex, involving many histopathological and molecular changes. Therefore, the characterization of cSCC will require additional investigations beyond genetic profiling. However, the analysis of gene expression may serve as a predictive tool in tumor development and progression, and may aid in identifying effective therapies in cSCC.
In summary, our findings have led to a better understanding of the mechanism, potential MFs and characteristics of cSCC via gene expression profiling and bioinformatics analysis. These candidate genes can serve as biomarkers for the diagnosis, treatment and evaluation of cSCC.
Conclusions
It is worth noting that the samples analyzed in this study originated from highly heterogeneous patient populations, which may have affected the gene expression profiles. Nonetheless, 335 candidate DEGs were obtained by GEO datasets and bioinformatics analysis. A total of 17 hub genes were found to be mainly related to the epithelial signal pathway, cell cycle and oocyte meiosis in cSCC and the high expression of CDK1 in CSCC samples was verified. Most importantly, these findings can improve our understanding of the causes and processes of cSCC. The identification of potential genes and pathways in CSCC could aid in identifying effective targeted therapies and contribute to its clinical management.
Acknowledgments
The authors thank Dr. Chaohu Wang for his expertise and guidance during the study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2945
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2945). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Landwehr MS, Watson SE, Macpherson CF, et al. The cost of cancer: a retrospective analysis of the financial impact of cancer on young adults. Cancer Med 2016;5:863-70. [Crossref] [PubMed]
- Rogers HW, Weinstock MA, Feldman SR, et al. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA Dermatol 2015;151:1081-6. [Crossref] [PubMed]
- Stratigos A, Garbe C, Lebbe C, et al. Diagnosis and treatment of invasive squamous cell carcinoma of the skin: European consensus-based interdisciplinary guideline. Eur J Cancer 2015;51:1989-2007. [Crossref] [PubMed]
- Keeping S, Xu Y, Chen CI, et al. Comparative efficacy of cemiplimab versus other systemic treatments for advanced cutaneous squamous cell carcinoma. Future Oncol 2020; [Epub ahead of print]. [Crossref] [PubMed]
- Sordillo JE, Kraft P, Wu AC, et al. Quantifying the Polygenic Contribution to Cutaneous Squamous Cell Carcinoma Risk. J Invest Dermatol 2018;138:1507-10. [Crossref] [PubMed]
- Chen X, Zhang FH, Chen QE, et al. The clinical significance of CDK1 expression in oral squamous cell carcinoma. Med Oral Patol Oral Cir Bucal 2015;20:e7-12. [Crossref] [PubMed]
- Bonelli P, Tuccillo FM, Borrelli A, et al. CDK/CCN and CDKI alterations for cancer prognosis and therapeutic predictivity. Biomed Res Int 2014;2014:361020. [Crossref] [PubMed]
- Zheng ZG, Ma BQ, Xiao Y, et al. Identification of biomarkers for the diagnosis and treatment of primary colorectal cancer based on microarray technology. Transl Cancer Res 2020;9:3453-67. [Crossref]
- Malumbres M. Physiological relevance of cell cycle kinases. Physiol Rev 2011;91:973-1007. [Crossref] [PubMed]
- Shintani S, Mihara M, Nakahara Y, et al. Expression of cell cycle control proteins in normal epithelium, premalignant and malignant lesions of oral cavity. Oral Oncol 2002;38:235-43. [Crossref] [PubMed]
- Wei W, Chen Y, Xu J, et al. Identification of Biomarker for Cutaneous Squamous Cell Carcinoma Using Microarray Data Analysis. J Cancer 2018;9:400-6. [Crossref] [PubMed]
- Feng H, Gu ZY, Li Q, et al. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J Ovarian Res 2019;12:35. [Crossref] [PubMed]
- Chang JT, Wang HM, Chang KW, et al. Identification of differentially expressed genes in oral squamous cell carcinoma (OSCC): overexpression of NPM, CDK1 and NDRG1 and underexpression of CHES1. Int J Cancer 2005;114:942-9. [Crossref] [PubMed]
- Zhao J, Han SX, Ma JL, et al. The role of CDK1 in apoptin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep 2013;30:253-9. [Crossref] [PubMed]
- Chae SW, Sohn JH, Kim DH, et al. Overexpressions of Cyclin B1, cdc2, p16 and p53 in human breast cancer: the clinicopathologic correlations and prognostic implications. Yonsei Med J 2011;52:445-53. [Crossref] [PubMed]
- Xi Q, Huang M, Wang Y, et al. The expression of CDK1 is associated with proliferation and can be a prognostic factor in epithelial ovarian cancer. Tumour Biol 2015;36:4939-48. [Crossref] [PubMed]
- Liu Y, Zhu YH, Mao CQ, et al. Triple negative breast cancer therapy with CDK1 siRNA delivered by cationic lipid assisted PEG-PLA nanoparticles. J Control Release 2014;192:114-21. [Crossref] [PubMed]
- Linton A, Cheng YY, Griggs K, et al. An RNAi-based screen reveals PLK1, CDK1 and NDC80 as potential therapeutic targets in malignant pleural mesothelioma. Br J Cancer 2018;118:e13. [Crossref] [PubMed]
- Cheng Y, Li K, Diao D, et al. Expression of KIAA0101 protein is associated with poor survival of esophageal cancer patients and resistance to cisplatin treatment in vitro. Lab Invest 2013;93:1276-87. [Crossref] [PubMed]
- Shi YX, Zhu T, Zou T, et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget 2016;7:85235-43. [Crossref] [PubMed]
- Yuan B, Xu Y, Woo JH, et al. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res 2006;12:405-10. [Crossref] [PubMed]
- Kim HS, Park KH, Kim SA, et al. Frequent mutations of human Mad2, but not Bub1, in gastric cancers cause defective mitotic spindle checkpoint. Mutat Res 2005;578:187-201. [Crossref] [PubMed]
- Hartwell LH, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci U S A 1970;66:352-9. [Crossref] [PubMed]
- Wan L, Tan M, Yang J, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell 2014;29:377-91. [Crossref] [PubMed]
- Taniguchi K, Momiyama N, Ueda M, et al. Targeting of CDC20 via small interfering RNA causes enhancement of the cytotoxicity of chemoradiation. Anticancer Res 2008;28:1559-63. [PubMed]
- Li J, Gao JZ, Du JL, et al. Increased CDC20 expression is associated with development and progression of hepatocellular carcinoma. Int J Oncol 2014;45:1547-55. [Crossref] [PubMed]
- Kim Y, Choi JW, Lee JH, et al. MAD2 and CDC20 are upregulated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the uterine cervix. Int J Gynecol Pathol 2014;33:517-23. [Crossref] [PubMed]
- Ding ZY, Wu HR, Zhang JM, et al. Expression characteristics of CDC20 in gastric cancer and its correlation with poor prognosis. Int J Clin Exp Pathol 2014;7:722-7. [PubMed]
- Choi JW, Kim Y, Lee JH, et al. High expression of spindle assembly checkpoint proteins CDC20 and MAD2 is associated with poor prognosis in urothelial bladder cancer. Virchows Arch 2013;463:681-7. [Crossref] [PubMed]
- Kidokoro T, Tanikawa C, Furukawa Y, et al. CDC20, a potential cancer therapeutic target, is negatively regulated by p53. Oncogene 2008;27:1562-71. [Crossref] [PubMed]
- Guo Y, Kim C, Ahmad S, et al. CENP-E--dependent BubR1 autophosphorylation enhances chromosome alignment and the mitotic checkpoint. J Cell Biol 2012;198:205-17. [Crossref] [PubMed]
- Malureanu LA, Jeganathan KB, Hamada M, et al. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell 2009;16:118-31. [Crossref] [PubMed]
- Lee E, Pain M, Wang H, et al. Sensitivity to BUB1B Inhibition Defines an Alternative Classification of Glioblastoma. Cancer Res 2017;77:5518-29. [Crossref] [PubMed]
- Zhuang L, Yang Z, Meng Z. Upregulation of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in Tumor Tissues Predicted Worse Overall Survival and Disease-Free Survival in Hepatocellular Carcinoma Patients. Biomed Res Int 2018;2018:7897346. [Crossref] [PubMed]
- Ma Q, Liu Y, Shang L, et al. The FOXM1/BUB1B signaling pathway is essential for the tumorigenicity and radioresistance of glioblastoma. Oncol Rep 2017;38:3367-75. [Crossref] [PubMed]
(English Language Editor: E Tan)


