
The expression and prognostic value of RNA binding proteins in clear cell renal cell carcinoma
Introduction
Clear cell renal cell carcinoma (ccRCC), originates from renal tubular epithelial cells, which is the most common histologic subtype of renal cell carcinoma (RCC) with an incidence accounting for about 75% of all cases (1). In recent year, owing to lack of apparent symptoms at the early stage, most ccRCC patients reach advanced stage of the cancer at the first diagnosis and miss the chance of accepting the radical treatment (2). Although significant progress has been made in diagnostic techniques and targeted therapies, most patients’ prognosis remains poor, mainly due to distant metastases and disease recurrence (3,4). To date, causing by the high recurrence and incidence in the ccRCC disease, it is quite urgent to find effective targeted biomarkers for ccRCC patients. In recent years, there is a lot of molecular research concerning on early diagnosis and prognosis, however, the missed diagnosis rates still very high (5). Therefore, it is highly valuable to find novel therapeutic targets for ccRCC.
RNA binding proteins (RBPs) are usually considered as proteins that bind to various types of RNAs. To date, at least 1,500 experimentally verified RBP coding genes have been identified in the human genome, accounting for about 7.5% of all protein-coding genes (6). They can interact with their target RNAs or proteins to form ribonucleoprotein (RNP) complexes to show extensive capabilities, such as regulation of mRNA stability, RNA localization, export, processing, splicing, degradation, and translation, which finally affects the expression of all genes in the cell (7). These abilities mean that abnormal deregulated RBPs are closely associated with the occurrence and development of diverse complex human diseases. Genetic mutations in RBPs, such as TDP-43, FUS, ATXN2, TAF15, EWSR1, hnRNPA1, hnRNPA2/B1, MATR3, and TIA1 may lead to amyotrophic lateral sclerosis (ALS) pathogenesis (8). Furthermore, previous studies have indicated that RBPs such as SRSF1, HuR, Rbm38, and QKI are known for their role in the occurrence and development of cardiovascular diseases (9). Despite the fact that RBPs closely involved in the initiation and progress of different types of human diseases, the roles of RBPs in the progression of human cancers are still unclear.
In recent years, abnormally expressed RBPs have been found in several tumors, which affected both the modification of RNA and the translation of mRNA into protein, contributing to tumor initiation and progression. Among these RBPs, very few have been investigated in depth and reported playing vital roles in human tumors. For instance, HuR could regulate the stability of mRNA, thereby promoting the proliferation and metastasis of gastric cancer cells (10); AGO2 by upregulating oncogenic miR-19b biogenesis to facilitates the progression of lung cancer (11); a previous study has revealed that ESRP1 was significantly upregulated in ovarian cancer at both mRNA and protein levels, and permits isoform switching from mesenchymal to epithelial phenotype in ovarian cancer cells (12). However, little is known about the functional roles of most RBPs in human cancers; an integrated study of RBPs will be greatly appreciated understanding their roles in tumors.
Gene expression data and corresponding clinicopathological information of ccRCC, which was downloaded from The Cancer Genome Atlas (TCGA) database, was applied for further analysis. In this current study, we screened differentially expressed RBPs found among cancerous tissues and healthy renal controls via comprehensive bioinformatics methods. Additionally, we performed gene ontology (GO) and pathway enrichment analysis, a PPI network complex construction, a module analysis and a hub genes selection. Furthermore, survival analysis, ROC analysis and Cox analysis were carried out to determine the clinical significance of RBP candidates in ccRCC. Finally, several online databases were employed to further explore the role of RPB candidates in ccRCC.
In conclusion, filtered potential RBPs might be associated with the occurrence and progression of ccRCC and could be recognized as biomarkers for diagnosis and prognosis and the formulation of drug targets.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2393).
Methods
Data collection and identification of differentially expressed RBPs
TCGA (https://portal.gdc.cancer.gov/) (13) database is comprehensive and the data that it contains is free to the public. In our study, we downloaded the RNA sequencing dataset of 72 noncancerous samples and 539 KIRC samples. All tumorous samples with unavailable or unknown survival time data will be ignored and finally we obtained RNA sequencing profile of 72 normal renal specimens and 530 ccRCC specimens. Additionally, relevant clinicopathological information of these 530 ccRCC patients, including age, gender, histological grade, clinical stage, and TMN stage was recorded in Table 1. Subsequently, The edgeR package (www.bioconductor.org/packages/release/bioc/html/edgeR.html) (14) was utilized to screen the differentially expressed RBPs among ccRCC tissues and noncancerous tissues, RBPs with adjusted P<0.05 and |log2fold change (FC)|≥1 were selected to perform further analysis. A volcano plot was utilized to visualize the identified differentially expressed RBPs.
Table 1
| Clinical parameters | Variable | Total (n=530) | Percentages (%) |
|---|---|---|---|
| Age | ≤60 | 264 | 49.81 |
| >60 | 266 | 50.19 | |
| Gender | Female | 186 | 35.09 |
| Male | 344 | 64.91 | |
| Histological grade | G1 | 14 | 2.64 |
| G2 | 227 | 42.83 | |
| G3 | 206 | 38.87 | |
| G4 | 75 | 14.15 | |
| GX | 5 | 0.94 | |
| Unknow | 3 | 0.57 | |
| Clinical stage | Stage I | 265 | 50 |
| Stage II | 57 | 10.74 | |
| Stage III | 123 | 23.21 | |
| Stage IV | 82 | 15.48 | |
| Unknow | 3 | 0.57 | |
| T classification | T1 | 271 | 51.13 |
| T2 | 69 | 13.02 | |
| T3 | 179 | 33.77 | |
| T4 | 11 | 2.08 | |
| Distant metastasis | M0 | 420 | 79.25 |
| M1 | 78 | 14.72 | |
| MX | 30 | 5.66 | |
| Unknow | 2 | 0.37 | |
| Lymph nodes | N0 | 239 | 45.09 |
| N1 | 16 | 3.02 | |
| NX | 275 | 51.89 |
TCGA, The Cancer Genome Atlas.
GO and pathway enrichment analysis
In order to investigate the potential mechanisms of differentially expressed genes (DEGs), the ClusterProfiler package (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (15) built in R, which could automate biological terminology classification and gene cluster enrichment analysis processes, was utilized to analyze the identified RBPs enrichment of biological process (BP), cellular component (CC) and molecular function (MF), and the Kyoto Encyclopedia of Genes and Genomes (KEGG). In the current study, function and signal pathway enrichment analyses were conducted with the criterion both P and FDR<0.05.
PPI network construction and app analysis
The online resource Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/) (16) was utilized to obtain protein-protein interaction (PPI) information of DEGs with the combined score of >0.400 (medium confidence score). Then, the PPI network was further constructed and visualized through the Cytoscape software (Version 3.6.0, http://www.cytoscape.org/). Subsequently, the CytoHubba plugin (17) in Cytoscape was utilized to screen hub RBPs by Degree method. Finally, the Molecular Complex Detection (MCODE) plugin (18) built in Cytoscape software was utilized to explore the significant module in PPI network with the following settings: cut-off degree, 2; node score cut-off, 0.2; Haircut, true; Fluff, false; K-core, 2; maximum depth,100. The ClusterProfiler package further executed the function and signal pathway enrichment analysis of genes in the most significant module.
Survival analysis, ROC analysis and COX analysis of hub RBPs
To evaluate the prognostic value of all hub RBPs, we performed the overall survival analysis of each hub RBPs, 530 ccRCC patients with available OS time data in the TCGA dataset were sorted into high- and low-expression subgroups according to the median expression level. RBPs with a criterion P<0.05 were considered as real candidate hub RBPs. Then, we calculated the receiver operating characteristic (ROC) curve using Graphpad Prism software (Version 8.0, https://www.graphpad.com/scientific-software/prism/) to evaluate the ability to discriminate healthy renal tissue and ccRCC tissue. Univariate and multivariate cox analyses was conducted to select independently candidate RBPs associated with the overall survival of ccRCC patients.
Comprehensive analysis of candidate RBPs
The gene expression array datasets of ONCOMINE (https://www.oncomine.org) are an online cancer microarray database (19). In this study, the ONCOMINE database was utilized to analyze mRNA levels of candidate RBPs in different cancers. The mRNA expressions of each candidate RBPs in different cancer samples were compared with that in normal controls, using a students’ t-test to obtain a P value. The cut-off of P value and fold change were defined as 0.001 and 1.5, respectively. UALCAN (http://ualcan.path.uab.edu) is a user-friendly interactive web database for analyzing cancer data, which could provide proteomic expression analysis option using data from the Clinical Proteomic Tumour Analysis Consortium (CPTAC) (20). In this work, we investigate the proteomic expression of candidate RBPs from the CPTAC-KIRC dataset. GEPIA (http://gepia.cancer-pku.cn) is a newly developed interactive web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects (21). It provides many customizable functions, including tumor/normal differential expression analysis, clinicopathology analysis, survival analysis, similar gene detection, correlation analysis and dimensionality reduction analysis. In this study, GEPIA was utilized to determine the correlation between candidate RBPs expression and ccRCC pathological stages. The Kaplan-Meier plotter (http://www.kmplot.com/) is able to assess the effect of 54k genes (mRNA, miRNA, protein) on survival in multiple tumor types (22). Using the Kaplan Meier plotter database, we further evaluated the effect of candidate RBPs on the survival of ccRCC patients. Finally, the function and signal pathway enrichment analysis of candidate genes were further performed by the ClusterProfiler package.
Statistical analysis
R software (version 3.6.1) and Graphpad Prism software (Version 8.0) were used for statistical analysis. The Kaplan-Meier curve was used to analyze the association between the expression level of candidate RBPs and the overall survival, with statistical significance evaluated using the Log-rank test. ROC curve was employed to analyzed the expression level of candidate RBPs to discriminate ccRCC patients and obtain the area under the curve. ONCOMINE database was analyzed by the students’ t-test to validate the expression levels of candidate RBPs in ccRCC samples and normal controls. P <0 .05 was considered statistically significant.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of DEGs
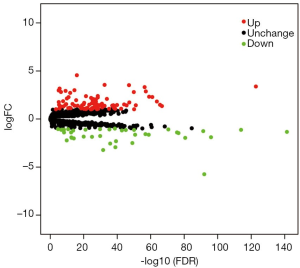
The edgeR package built in R software was used to identify the DEGs between ccRCC tissues and noncancerous tissues. A total of 1,542 RBPs were included in this study, and 133 RBPs, including 94 upregulated RBPs and 39 downregulated RBPs, were identified between ccRCC samples and noncancerous samples with the setting standard |log2FC| ≥1 & the adjusted P value <0.05 (Table 2). In addition, Figure 1 displayed the volcano map of all DEGs. These DEGs were chosen for further analysis.
Table 2
| DEGs | Gene symbol |
|---|---|
| Up-regulated | RPS20, DARS, ZFR2, RPS14, CLK2, NOL12, RPL35, THOC6, RPL28, NOP16, APOBEC3F, RPL18A, RPL13, SRRM3, RPL18, U2AF1L4, CLK1, TRMT1, EEF1G, TDRD10, NOVA2, QTRT1, ZC3HAV1L, MEX3B, AFF2, CLASRP, RPL36, ELAVL2, RPL36A, PABPC4L, PIWIL4, EXOSC5, OAS2, YBX2, RPL10L, RPL22L1, AEN, SAMHD1, RBM44, MOV10L1, OAS1, RPS2, AFF3, RPS19, GAPDH, CLK4, CELF6, TLR3, RTL3, YBX3, ZC3H12D, RNASE6, FBXO17, OASL, ELAVL3, DDX39B, U2AF1, DDX47, ARL6IP4, POLR2F, EXO1, ANG, ELAVL4, INTS6L, EEF1A2, TDRD6, EZH2, CELF5, RDM1, NXF5, TLR7, TLR8, EIF4A1, PIWIL3, PABPC1L, RNASE10, RNF113B, APOBEC3G, ISG20, RNASE3, RNASE2, DQX1, PATL2, DDX53, APOBEC3H, NANOS2, DAZ1, IGF2BP3, NR0B1, RNASET2, NOL3, JAKMIP1, RBM46, TERT |
| Down-regulated | RALYL, ESRP1, TDRD5, RBM11, DDX25, KHDRBS2, LIN28A, RBFOX1, ENOX1, TDRD1, DAZL, C2orf15, CELF3, AZGP1, PPARGC1A, MRPS6, NANOS1, ESRP2, LARS2, CSDC2, CPEB3, ADAD2, CNP, SNRPN, NXF3, AUH, ACO1, RBM47, MRPL33, TDRD9, IGF2BP2, TST, PIH1D3, MSI2, IPO13, CPEB4, NPM2, TDRD15, STRBP |
DEGs, differentially expressed genes.
GO and KEGG pathway enrichment analyses of DEGs
To further investigate the potential mechanisms of DEGs in ccRCC, we performed GO and pathway enrichment analyses of DEGs using the ClusterProfiler package in R software with the criterion adjusted P<0.05. As can be seen from Table 3, in the BP group, upregulated DEGs were enriched mainly in RNA catabolic process, mRNA catabolic process, RNA splicing, response to virus, and nuclear-transcribed mRNA catabolic process; downregulated DEGs were mainly involved in the regulation of mRNA metabolic process, cellular process involved in reproduction in multicellular organism, regulation of translation, RNA splicing, and regulation of cellular amide metabolic process. In the CC group, upregulated DEGs were mainly enriched in ribosome, cytosolic ribosome, ribosomal subunit, cytoplasmic ribonucleoprotein granule, and ribonucleoprotein granule; downregulated DEGs were mainly involved in cytoplasmic ribonucleoprotein granule, ribonucleoprotein granule, mitochondrial matrix, chromatoid body, and P granule. In the MF group, upregulated DEGs were mainly enriched in catalytic activity, acting on RNA, structural constituent of ribosome, nuclease activity, ribonuclease activity, and double-stranded RNA binding; downregulated DEGs were mainly involved in mRNA 3’-UTR binding, translation regulator activity, single-stranded RNA binding, translation repressor activity, translation regulator activity, and nucleic acid binding. According to the KEGG pathway enrichment analysis, all DEGs were enriched mainly in Ribosome, RNA transport, Legionellosis, mRNA surveillance pathway, Influenza A (Table 4).
Table 3
| Category | Terms | Description | FDR | Count |
|---|---|---|---|---|
| Up-regulated | ||||
| BP | GO:0006401 | RNA catabolic process | 3.00E-19 | 25 |
| BP | GO:0006402 | mRNA catabolic process | 9.71E-13 | 19 |
| BP | GO:0008380 | RNA splicing | 3.71E-10 | 18 |
| BP | GO:0009615 | response to virus | 1.26E-09 | 15 |
| BP | GO:0000956 | Nuclear-transcribed mRNA catabolic process | 1.43E-10 | 14 |
| CC | GO:0005840 | Ribosome | 4.75E-10 | 14 |
| CC | GO:0022626 | Cytosolic ribosome | 3.81E-13 | 13 |
| CC | GO:0044391 | Ribosomal subunit | 1.88E-10 | 13 |
| CC | GO:0036464 | Cytoplasmic ribonucleoprotein granule | 3.77E-10 | 13 |
| CC | GO:0035770 | Ribonucleoprotein granule | 4.75E-10 | 13 |
| MF | GO:0140098 | Catalytic activity, acting on RNA | 1.04E-11 | 19 |
| MF | GO:0003735 | Structural constituent of ribosome | 2.45E-09 | 13 |
| MF | GO:0004518 | Nuclease activity | 2.45E-07 | 11 |
| MF | GO:0004540 | Ribonuclease activity | 2.45E-07 | 9 |
| MF | GO:0003725 | Double-stranded RNA binding | 2.36E-07 | 8 |
| Down-regulated | ||||
| BP | GO:1903311 | Regulation of mRNA metabolic process | 5.65E-06 | 9 |
| BP | GO:0022412 | Cellular process involved in reproduction in multicellular organism | 7.41E-05 | 8 |
| BP | GO:0006417 | Regulation of translation | 1.63E-04 | 8 |
| BP | GO:0008380 | RNA splicing | 2.88E-04 | 8 |
| BP | GO:0034248 | Regulation of cellular amide metabolic process | 3.07E-04 | 8 |
| CC | GO:0036464 | Cytoplasmic ribonucleoprotein granule | 6.50E-04 | 5 |
| CC | GO:0035770 | Ribonucleoprotein granule | 6.89E-04 | 5 |
| CC | GO:0005759 | Mitochondrial matrix | 1.39E-02 | 5 |
| CC | GO:0033391 | Chromatoid body | 1.22E-04 | 3 |
| CC | GO:0043186 | P granule | 1.22E-04 | 3 |
| MF | GO:0003730 | mRNA 3'-UTR binding | 2.78E-06 | 6 |
| MF | GO:0045182 | Translation regulator activity | 6.59E-06 | 5 |
| MF | GO:0003727 | Single-stranded RNA binding | 5.58E-04 | 4 |
| MF | GO:0030371 | Translation repressor activity | 5.58E-04 | 3 |
| MF | GO:0090079 | Translation regulator activity, nucleic acid binding | 5.58E-04 | 3 |
GO, gene ontology; FDR, false discovery rate; BP, biological process; CC, cellular components; MF, molecular function.
Table 4
| Category | Terms | Description | FDR | Count |
|---|---|---|---|---|
| KEGG | hsa03010 | Ribosome | 4.89E-11 | 13 |
| KEGG | hsa03013 | RNA transport | 9.57E-04 | 7 |
| KEGG | hsa05134 | Legionellosis | 3.33E-03 | 4 |
| KEGG | hsa03015 | mRNA surveillance pathway | 1.48E-02 | 4 |
| KEGG | hsa05164 | Influenza A | 1.78E-02 | 5 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate.
PPI network construction and app analysis
A total of 133 identified RBPs were filtered for the STRING website and further analyzed through Cytoscape software, and we constructed a PPI network with 111 nodes and 413 edges (Figure 2A). Subsequently, ten hub RBPs with the highest degree, including RPS14, RPS2, GAPDH, RPS20, RPL35, EIF4A1, RPL18, RPL13, RPL18A, and RPS19, were selected for further analysis by cytohubba plugin. The most significant module was selected from PPI network through the MCODE plugin built into the Cytoscape software (Figure 2B). This significant module was consisted of 17 upregulated RBPs/nodes and 131 edges, which enriched particularly in translational initiation, SRP-dependent cotranslational protein targeting to membrane, cotranslational protein targeting to membrane, protein targeting to ER, nuclear-transcribed mRNA catabolic process, and nonsense-mediated decay (BP); cytosolic ribosome, ribosomal subunit, cytosolic part, ribosome, and cytosolic large ribosomal subunit (CC); structural constituent of ribosome, translation factor activity, RNA binding, translation elongation factor activity, fibroblast growth factor binding, and double-stranded RNA binding (MF); and Ribosome, and Legionellosis (KEGG) (Table 5).
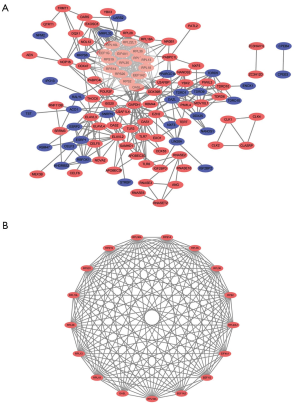
Table 5
| Category | Terms | Description | P.adjust | Count |
|---|---|---|---|---|
| BP | GO:0006413 | Translational initiation | 1.31E-19 | 12 |
| BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 1.08E-19 | 11 |
| BP | GO:0006613 | Cotranslational protein targeting to membrane | 1.08E-19 | 11 |
| BP | GO:0045047 | Protein targeting to ER | 1.31E-19 | 11 |
| BP | GO:0000184 | Nuclear-transcribed mRNA catabolic process, nonsense-mediated decay | 1.31E-19 | 11 |
| CC | GO:0022626 | Cytosolic ribosome | 1.90E-25 | 13 |
| CC | GO:0044391 | Ribosomal subunit | 1.22E-22 | 13 |
| CC | GO:0044445 | Cytosolic part | 2.69E-21 | 13 |
| CC | GO:0005840 | Ribosome | 7.24E-21 | 13 |
| CC | GO:0022625 | Cytosolic large ribosomal subunit | 2.37E-18 | 9 |
| MF | GO:0003735 | Structural constituent of ribosome | 2.42E-21 | 13 |
| MF | GO:0008135 | Translation factor activity, RNA binding | 9.70E-04 | 3 |
| MF | GO:0003746 | Translation elongation factor activity | 1.52E-03 | 2 |
| MF | GO:0017134 | Fibroblast growth factor binding | 1.52E-03 | 2 |
| MF | GO:0003725 | Double-stranded RNA binding | 1.30E-02 | 2 |
| KEGG | hsa03010 | Ribosome | 4.43E-19 | 13 |
| KEGG | hsa05134 | Legionellosis | 1.57E-02 | 2 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; BP, biological process; CC, cellular components; MF, molecular function.
Survival analysis, ROC analysis and Cox analysis of hub RBPs
To explore the prognostic value of hub RBPs, Kaplan-Meier curve for overall survival of ccRCC patients was calculated according to the low and high expressions of each hub RBP. As shown in Figure 3, eight candidate hub RBPs, including RPS2 (P=0.001), GAPDH (P=0.014), RPS20 (P=2.396E-04), EIF4A1 (P=3.817E-07), RPL18 (P=0.002), RPL13 (P=8.9E-04), RPL18A (P=0.022), and RPS19 (P=4.672E-05) have lower percent survival in the high-expression group compared to the low-expression group. To evaluate the diagnostic value of these eight hub RBPs further, the receiver operating characteristic (ROC) curve was calculated to assess the ability to distinguish ccRCC tissue from noncancerous renal tissue. As shown in Figure 4, the area under the curve (AUC) of hub RBPs RPS2 (AUC = 0.95, P<0.0001), GAPDH (AUC = 0.96, P<0.0001), RPS20 (AUC = 0.88, P<0.0001), EIF4A1 (AUC = 0.89, P<0.0001), RPL18 (AUC = 0.91, P<0.0001), RPL13 (AUC = 0.89, P<0.0001), RPL18A (AUC = 0.90, P<0.0001), and RPS19 (AUC = 0.94, P<0.0001), indicates that the eight candidate hub RBPs have better diagnostic efficiency for ccRCC. Moreover, as shown in Table 6, in Univariate analysis, we found that high RPS20 expression (HR, 1.568, P=2.30E-03), high RPL18 expression (HR, 1.504, P=7.12E-03), high RPS19 expression (HR, 1.578, P=9.81E-04), high RPS2 expression (HR, 1.630, P=1.44E-03), high EIF4A1 expression (HR, 1.281, P=1.63E-03), high RPL13 expression (HR, 1.514, P=4.16E-03), age (HR, 1.023; P=0.012), histological grade (HR, 2.242; P=3.61E-08), clinical stage (HR, 1.862; P=1.26E-10), T classification (HR, 1.943; P=2.69E-08), Lymph nodes (HR, 2.932; P=0.001) and distant metastasis (HR, 4.073; P=2.76E-10) were associated with poorer overall survival of patients with ccRCC. Furthermore, multivariate survival model after variable selection indicated that age (HR, 1.035; P=1.29E-04), histological grade (HR, 1.471, P=4.19E-02), increased EIF4A1 expression (HR, 1.284; P=9.01E-04) and high RPL13 expression of patients with ccRCC (HR, 1.503; P=5.73E-03) were independently associated with unfavorable OS. The above results showed that EIF4A1 or RPL13 could serve as an individual predictor for poor prognosis in ccRCC.
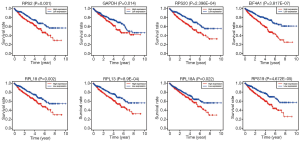
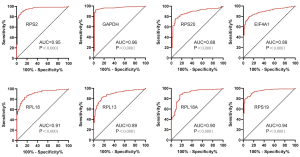
Table 6
| Clinicopathological variables | Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age | 2.500 (1.1743–5.322) | 1.75E-02 | 1.030 (1.019–1.061) | 1.29E-04 | |
| Gender | 1.013 (0.666–1.541) | 9.51E-01 | |||
| Histological grade | 2.242 (1.682–2.988) | 3.61E-08 | 1.471 (1.014–2.135) | 4.19E-02 | |
| Clinical stage | 1.862 (1.541–2.251) | 1.26E-10 | 1.605 (0.911–2.825) | 1.01E-01 | |
| T classification | 1.943 (1.538–2.456) | 2.69E-08 | 0.917 (0.545–1.544) | 7.45E-01 | |
| Distant metastasis | 4.073 (2.634–6.300) | 2.76E-10 | 2.018 (0.792–5.141) | 1.41E-01 | |
| Lymph nodes | 2.932 (1.516–5.668) | 1.00E-03 | 1.828 (0.845–3.956) | 1.26E-01 | |
| RPS20 | 1.568 (1.174–2.093) | 2.30E-03 | 1.182 (0.688–2.032) | 5.45E-01 | |
| RPL18 | 1.504 (1.117–2.024) | 7.12E-03 | 0.820 (0.480–1.400) | 4.67E-01 | |
| RPS19 | 1.578 (1.203–2.069) | 9.81E-04 | 1.269 (0.765–2.106) | 3.57E-01 | |
| RPL18A | 1.311 (0.957–1.796) | 9.21E-02 | |||
| GAPDH | 1.202 (0.910–1.587) | 1.94E-01 | |||
| RPS2 | 1.630 (1.207–2.200) | 1.44E-03 | 1.411 (0.858–2.323) | 1.75E-01 | |
| EIF4A1 | 1.281 (1.098–1.495) | 1.63E-03 | 1.284 (1.108–1.489) | 9.01E-04 | |
| RPL13 | 1.514 (1.140–2.012) | 4.16E-03 | 1.503 (1.126–2.008) | 5.73E-03 | |
ccRCC, clear cell renal cell carcinoma; TCGA, The Cancer Genome Atlas, HR, Hazard ratio.
Comprehensive analysis of candidate RBPs
Using ONCOMINE database, we compare the mRNA expression of the above eight candidate RBPs within cancer specimens and healthy control samples (Figure 5 and Table 7). Results showed that the mRNA expression of RPS2, GAPDH, EIF4A1, RPL18, RPL13, RPL18A, and RPS19 was overexpressed in ccRCC patients. Although the mRNA expression of RPS20 was also slightly upregulated in ccRCC samples than in normal renal samples with a P value of <0.001, the cut-off of fold change was <1.5. Additionally, to determine the proteomic expression of eight hub RBPs in ccRCC, we used protein expression data from the CPTAC-KIRC dataset to show that RPS20 (P=4.71E-07), GAPDH (P=4.85E-59), EIF4A1 (P=5.00E-39), RPL18 (P=9.46E-23), RPL13 (P=7.87E-35), RPL18A (P=1.14E-15), and RPS19 (P=2.25E-15) were significantly upregulated in ccRCC tissues by comparison with normal renal tissues. However, the RPS2 (P=2.22E-03) protein expression was relatively decreased in ccRCC tissues (Figure 6). Furthermore, using the GEPIA database, we also analyzed the expression of eight candidate RBPs with tumor stage. Results showed that RPS2 (P=5.08E-05), GAPDH (P=8.28E-04), RPS20 (P=8.54E-06), RPL13 (P=1.97E-03), and RPS19 (P=2.16E-04) groups significantly varied, whereas EIF4A1 (P=2.17E-01), RPL18 (2.98E-01), and RPL18A (P=4.72E-01) groups did not significantly differ (Figure 7). Furthermore, we employed the Kaplan-Meier Plotter database to assess the prognostic value of candidate RBPs in ccRCC patients. As shown in Figure 8, we found that higher expression of RPS2 (HR, 1.84, P=7E-05), GAPDH (HR, 1.82, P=1.3E-04), RPS20 (HR, 2.05, P=1.4E-06), EIF4A1 (HR, 2.19, P=2.2E-07), RPL18 (HR, 1.65, P=1.1E-03), RPL13 (HR, 1.95, P=4.4E-05), RPL18A (HR, 1.339, P=0.12), or RPS19 (HR, 2.01, P=1.2E-05) indicates a worse prognosis for ccRCC patients, however, there is no statistical difference in RPL18A. Finally, we performed GO and KEGG pathway analyses of eight RBPs. The results showed that these hub RBPs are mainly associated with translational initiation (BP), cytosolic ribosome (CC), structural constituent of ribosome (MF), and Ribosome (KEGG) (Table 8).
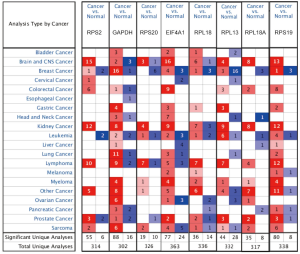
Table 7
| Gene | Types of ccRCC vs. normal samples | Fold change | P value | t-test | Reference |
|---|---|---|---|---|---|
| RPS2 | Clear cell renal cell carcinoma vs. normal | 1.806 | 1.61E-05 | 6.159 | Lenburg Renal (23) |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.601 | 2.30E-09 | 8.773 | Beroukhim Renal (24) | |
| Hereditary clear cell renal cell carcinoma vs. normal | 1.726 | 3.62E-09 | 12.244 | Beroukhim Renal (24) | |
| Clear cell renal cell carcinoma vs. normal | 2.072 | 2.09E-06 | 6.864 | Gumz Renal (25) | |
| Clear cell renal cell carcinoma vs. normal | 1.910 | 1.38E-13 | 11.405 | Jones Renal (26) | |
| GAPDH | Clear cell renal cell carcinoma vs. normal | 1.566 | 7.04E-06 | 8.317 | Yusenko Renal (27) |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.594 | 2.28E-06 | 6.755 | Beroukhim Renal (24) | |
| RPS20 | Clear cell renal cell carcinoma vs. normal | 1.489 | 2.18E-12 | 9.471 | Jones Renal (26) |
| EIF4A1 | Clear cell renal cell carcinoma vs. normal | 1.915 | 4.09E-10 | 9.519 | Higgins Renal (28) |
| Clear cell renal cell carcinoma vs. normal | 1.702 | 7.40E-08 | 8.289 | Gumz Renal (25) | |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.918 | 1.75E-07 | 7.381 | Beroukhim Renal (24) | |
| Hereditary clear cell renal cell carcinoma vs. normal | 2.172 | 1.16E-08 | 9.959 | Beroukhim Renal (24) | |
| Clear cell renal cell carcinoma vs. normal | 2.416 | 6.33E-10 | 7.945 | Jones Renal (26) | |
| RPL18 | Clear cell renal cell carcinoma vs. normal | 1.724 | 1.11E-13 | 10.468 | Jones Renal (26) |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.538 | 1.19E-06 | 6.081 | Beroukhim Renal (24) | |
| Hereditary clear cell renal cell carcinoma vs. normal | 1.662 | 1.83E-07 | 8.455 | Beroukhim Renal (24) | |
| RPL13 | Clear cell renal cell carcinoma vs. normal | 1.571 | 3.51E-05 | 5.456 | Lenburg Renal (23) |
| Clear cell renal cell carcinoma vs. normal | 1.650 | 6.95E-16 | 12.105 | Jones Renal (26) | |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.500 | 4.38E-08 | 6.951 | Beroukhim Renal (24) | |
| Hereditary clear cell renal cell carcinoma vs. normal | 1.508 | 3.76E-08 | 9.046 | Beroukhim Renal (24) | |
| Clear cell renal cell carcinoma vs. normal | 2.240 | 9.40E-05 | 5.453 | Gumz Renal (25) | |
| RPL18A | Clear cell renal cell carcinoma vs. normal | 1.989 | 4.32E-11 | 11.592 | Higgins Renal (28) |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.847 | 3.17E-09 | 8.684 | Beroukhim Renal (24) | |
| Hereditary clear cell renal cell carcinoma vs. normal | 2.212 | 1.84E-09 | 13.662 | Beroukhim Renal (24) | |
| Clear cell renal cell carcinoma vs. normal | 2.330 | 3.65E-05 | 5.574 | Gumz Renal (25) | |
| Clear cell renal cell carcinoma vs. normal | 2.246 | 9.35E-04 | 4.225 | Lenburg Renal (23) | |
| RPS19 | Clear cell renal cell carcinoma vs. normal | 2.123 | 5.71E-06 | 6.447 | Lenburg Renal (23) |
| Non-hereditary clear cell renal cell carcinoma vs. normal | 1.889 | 7.32E-11 | 9.763 | Beroukhim Renal (24) | |
| Hereditary clear cell renal cell carcinoma vs. normal | 2.024 | 1.58E-10 | 13.236 | Beroukhim Renal (24) | |
| Clear cell renal cell carcinoma vs. normal | 2.410 | 5.75E-05 | 8.001 | Higgins Renal (28) | |
| Clear cell renal cell carcinoma vs. normal | 2.303 | 5.41E-06 | 6.347 | Gumz Renal (25) | |
| Clear cell renal cell carcinoma vs. normal | 2.887 | 2.25E-10 | 8.498 | Jones Renal (26) |
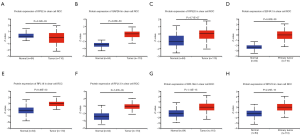
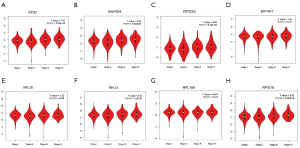
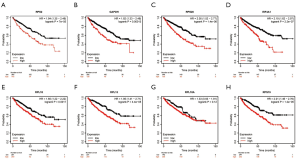
Table 8
| Category | Terms | Description | FDR | Count |
|---|---|---|---|---|
| BP | GO:0006413 | translational initiation | 1.82E-11 | 7 |
| BP | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane | 6.45E-11 | 6 |
| BP | GO:0006613 | cotranslational protein targeting to membrane | 6.45E-11 | 6 |
| CC | GO:0022626 | cytosolic ribosome | 1.79E-11 | 6 |
| CC | GO:0044391 | ribosomal subunit | 2.24E-10 | 6 |
| CC | GO:0044445 | cytosolic part | 7.31E-10 | 6 |
| MF | GO:0003735 | structural constituent of ribosome | 1.47E-09 | 6 |
| MF | GO:0017134 | fibroblast growth factor binding | 5.85E-04 | 2 |
| MF | GO:0019838 | growth factor binding | 1.40E-02 | 2 |
| KEGG | hsa03010 | Ribosome | 4.19E-08 | 6 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; BP, biological process; CC, cellular components; MF, molecular function.
Discussion
ccRCC is a heterogeneous disease with complicated pathogenesis, and an unclear underlying action mechanism. A large number of evidence showed that most ccRCC are characterized by dysregulation of hypoxia-inducible factor signaling, mutations in several vital histone and chromatin modifying enzyme and metabolic reprogramming in cell metabolism (29,30). Although diagnostic techniques and targeted therapies have been developed over the past few decades, the prognosis of this cancer remained unsatisfactory. Therefore, screening novel biomarkers for initial diagnosis and therapy of ccRCC is of great significance. High-throughput sequencing technology and bioinformatics have been rapidly developed, which has laid the foundation for discovering new molecular targets for diagnosis, treatment of cancers and prediction of their prognosis.
In recent years, many studies have demonstrated that RBPs play a crucial role in the initiation and progression of different types of human cancers. However, only a very few of RBPs have been examined in depth. The specific functional role of most RBPs in the process of tumorigenesis remains relatively unexplored. In the current study, we aimed to explore the potential diagnostic and prognostic RBPs for ccRCC patients through an integrated bioinformatics analysis.
Here, a total of 133 differentially expressed RBPs, consisting of 94 upregulated genes and 39 down-regulated genes, were screened between ccRCC samples and noncancerous samples. We then investigated relevant functional pathways of these differentially expressed RBPs, which enhanced our understanding of the pathogenesis of ccRCC. Subsequently, a protein interaction network of these differentially expressed RBPs was constructed. Subsequently, ten hub genes were screened from the PPI network, the survival analysis showed that evaluated expression of these eight candidate RBPs, including RPS2, GAPDH, RPS20, EIF4A1, RPL18, RPL13, RPL18A, and RPS19, is significantly correlated with poor overall survival of ccRCC patients, further ROC analysis found that these eight RBPs have considerable diagnostic accuracy to distinguish patients with ccRCC from healthy control subjects. In addition, cox regression analysis showed that most of candidate RBPs were associated with overall survival, and EIF4A1 and RPL13 could serve as independent risk factor to predict the overall survival of ccRCC. Then, to investigate the expression levels of these eight genes further, we used the ONCOMINE database to examine the mRNA expression, the results revealed that eight genes were differently upregulated in ccRCC specimens compare with in normal renal samples. Additionally, proteomic expression analysis showed that almost all key RBPs were high expressed in ccRCC samples compared to normal renal samples, except for RPS2. Furthermore, clinicopathological and survival analysis indicated that increased expression of most of candidate RBPs was associated with advanced stage and worse prognosis.
Among these eight genes, GAPDH is proven to be overexpressed in bone metastatic primary renal cell carcinoma at protein levels, and elevated expression predicted adverse prognosis (31). Even though the relationship between other candidate RBPs and ccRCC remains unclear, some of them have been demonstrated to be closely correlated with other cancers. For instance, RPS2 is overexpressed in prostate cancer, and knockdown of RBPS2 results in growth inhibition and apoptosis of tumor cells (32). RPS20 is significantly increased in glioma and negatively related to patients’ prognosis (33). EIF4A1 facilitates epithelial-mesenchymal transition and metastasis of gastric cancer (34). RPL13 is significantly increased in freshly resected cancer tissue of the stomach, colorectum, and liver, and knocking down RPL13 expression results in the drastic suppression of tumor cell growth with significant G1 and G2/M arrest of the cell cycle (35). Finally, by performing GO and KEGG pathway analysis of eight candidate RBPs, we inferred that eight candidate RBPs might lead to ccRCC by affecting translational initiation, cytosolic ribosome, structural constituent of ribosome, and Ribosome. These findings suggested that candidate hub genes played a specific function in the progression of ccRCC.
In conclusion, our study concluded that RPS2, GAPDH, RPS20, EIF4A1, RPL18, RPL13, RPL18A, and RPS19 might serve as the potential diagnostic and prognostic markers for ccRCC patients. However, this conclusion has yet to be verified in future studies, clinical samples are needed for further molecular and functional studies. Anyway, we hope the conclusion may provide some useful information and directions for the promising biomarkers and action mechanisms of ccRCC.
Acknowledgments
The authors thank Bo Zhao (Official Wechat Account: SCIPhD) of ShengXinZhuShou for suggestions on the manuscript.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2393
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2393). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The public databases mentioned in this study are publicly available for re-analyzing, and no ethical approval was required by the local ethics committees, so that this study does not require the ethics approval.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Barata PC, Rini BI. Treatment of renal cell carcinoma: Current status and future directions. CA Cancer J Clin 2017;67:507-24. [Crossref] [PubMed]
- Beroukhim R, Brunet JP, Di Napoli A, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res 2009;69:4674-81. [Crossref] [PubMed]
- Chandrashekar D S, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Chen W, Hill H, Christie A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016;539:112-7. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- de Bruin RG, Rabelink TJ, van Zonneveld AJ, et al. Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur Heart J 2017;38:1380-8. [Crossref] [PubMed]
- Fernández-Pello S, Hofmann F, Tahbaz R, et al. A Systematic Review and Meta-analysis Comparing the Effectiveness and Adverse Effects of Different Systemic Treatments for Non-clear Cell Renal Cell Carcinoma. Eur Urol 2017;71:426-36. [Crossref] [PubMed]
- Gao C, Guo X, Xue A, et al. High intratumoral expression of eIF4A1 promotes epithelial-to-mesenchymal transition and predicts unfavorable prognosis in gastric cancer. Acta Biochim Biophys Sin (Shanghai) 2020;52:310-9. [Crossref] [PubMed]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet 2014;15:829-45. [Crossref] [PubMed]
- Gumz ML, Zou H, Kreinest PA, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res 2007;13:4740-9. [Crossref] [PubMed]
- Hentze MW, Castello A, Schwarzl T, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 2018;19:327-41. [Crossref] [PubMed]
- Higgins JP, Shinghal R, Gill H, et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol 2003;162:925-32. [Crossref] [PubMed]
- Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers 2017;3:17009. [Crossref] [PubMed]
- Jeong HM, Han J, Lee SH, et al. ESRP1 is overexpressed in ovarian cancer and promotes switching from mesenchymal to epithelial phenotype in ovarian cancer cells. Oncogenesis 2017;6:e389. [Crossref] [PubMed]
- Jones J, Otu H, Spentzos D, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res 2005;11:5730-9. [Crossref] [PubMed]
- Kobayashi T, Sasaki Y, Oshima Y, et al. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med 2006;18:161-70. [Crossref] [PubMed]
- Lenburg ME, Liou LS, Gerry NP, et al. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer 2003;3:31. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Rhodes D R, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1-6. [Crossref] [PubMed]
- Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010;26:139-40. [Crossref] [PubMed]
- Sanchez DJ, Simon MC. Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochim Biophys Acta Rev Cancer 2018;1870:23-31. [Crossref] [PubMed]
- Szendrői A, Szász AM, Kardos M, et al. Opposite prognostic roles of HIF1α and HIF2α expressions in bone metastatic clear cell renal cell cancer. Oncotarget 2016;7:42086-98. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-102. [Crossref] [PubMed]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68-77. [Crossref] [PubMed]
- Vera-Badillo FE, Templeton AJ, Duran I, et al. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur Urol 2015;67:740-9. [Crossref] [PubMed]
- Wang M, Hu Y, Stearns ME. RPS2: a novel therapeutic target in prostate cancer. J Exp Clin Cancer Res 2009;28:6. [Crossref] [PubMed]
- Wettersten HI, Aboud OA, Lara PN Jr, et al. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol 2017;13:410-9. [Crossref] [PubMed]
- Xie M, Ma T, Xue J, et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett 2019;443:67-79. [Crossref] [PubMed]
- Yong WH, Shabihkhani M, Telesca D, et al. Ribosomal Proteins RPS11 and RPS20, Two Stress-Response Markers of Glioblastoma Stem Cells, Are Novel Predictors of Poor Prognosis in Glioblastoma Patients. PloS One 2015;10:e0141334. [Crossref] [PubMed]
- Yu G, Wang L G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Yusenko MV, Kuiper RP, Boethe T, et al. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer 2009;9:152. [Crossref] [PubMed]
- Zhang H, Wang Y, Dou J, et al. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene 2019;38:1410-31. [Crossref] [PubMed]
- Zhao M, Kim JR, Van Bruggen R, et al. RNA-Binding Proteins in Amyotrophic Lateral Sclerosis. Mol Cells 2018;41:818-29. [PubMed]


