
MicroRNA-133b expression inversely correlates with MET and can serve as an optimum predictive biomarker for patients of colorectal cancer
Introduction
Colorectal cancer (CRC) is a leading cause of cancer death worldwide, with over 1.2 million new cases and a death toll of over 600,000 annually (1). The rate of incidence and mortality still maintains a steep increase in China, only in 2011, both peaked at 23.03/100,000 and 11.11/100,000, respectively, only inferior to lung cancer and gastric cancer (2). Two-thirds of CRC patients in China have already reached advanced disease stages upon diagnosis. The current situation is that surgery is the primary treatment for CRC, and the 5-year survival rates is low, ranging from 30% to 70%, which is especially true for the patients at advanced stages (3-5). Hence, unraveling the mechanisms through which CRC cells drive tumor progression and identifying novel, sensitive and specific diagnostic biomarkers may allow clinicians to increase diagnostic efficiency and ultimately improved CRC treatment.
MicroRNAs (miRNAs) are small, non-coding RNAs that modulate gene expression by inhibiting or inactivating target messenger RNAs (mRNAs) (6). An increasing amount of evidence indicates that the altered expression of miRNAs is strongly implicated in tumorigenesis and progression of various malignancies. By impairing the functions of their corresponding target genes, miRNAs work as either oncogenes or tumor suppressors and regulate various cancer-related signaling pathways. Furthermore, it has been indicated that these abnormal miRNAs may possess the potential to be used as biomarkers for human malignancies and to diagnose cancers or prognosticate their advancement (7-10).
As an important tumor suppressor, previously studies have implied that microRNA-133b (miR-133b) significantly downregulated in various malignancies and can serve as a tumor suppressor. Notably, miR-133b inhibits the tumor migration and invasion via targeting FSCN1 in gastric cancer and lung cancer, via targeting matrix metalloproteinase (MMP-9) in renal cancer and via targeting MMP-14 in hepatocarcinoma and glioblastoma (11-15). We had previously analyzed the role of miR-133b involved in CRC and found that the expression of it is greatly downregulated in human CRC cells, and importantly, re-expression of miR-133b depress the growth of CRC cells in vitro and in vivo by targeting mesenchymal-epithelial transition factor (MET) (16).
MET, the receptor tyrosine kinase for hepatocyte growth factors (HGFs), is a membrane receptor for the HGF; It is well known that MET tyrosine kinase can increase the survival rate of many cells when exposed to various apoptotic stimulants such as serum starvation, death receptor activation, or genotoxic treatment (17,18). A large body of evidence has shown that MET participates in the occurrence and progression of tumors. Therefore, MET is considered an important target of anti-cancer therapy, and MET signaling inhibitors such as ligand antagonists, kinase inhibitors and receptor competitors have been developed (19). However, the relationship between miR-133b and MET expression level remains obscure in CRC tissues. Furthermore, much still remains unknown about the correlation between miR-133b expression and clinicopathological parameters. The purpose of this study was to validate the role of miR-133b as a diagnostic marker and to determine its association with MET and clinicopathological parameters in CRC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2370).
Methods
Clinical materials
Biological samples (46 pairs CRC tissues and non-cancerous tissues) used in this article were obtained from patients without receiving radiotherapy or chemotherapy prior to surgery treated at the Third Xiangya Hospital of Central South University (Hunan, China) under informed consent and approved by the Ethics Committee of Central South University (approval number: No. 2016-s104). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The histopathological type and stage of CRC were evaluated according to the criteria of the World Health Organization classification system. All patients were separated according to their stages using the US National Comprehensive Cancer Network (NCCN) Clinical Guidelines 2016.
RNA extraction and quantitative real-time RT-PCR
miR-133b and total RNA of MET was extracted from tissues using EZNA PF miRNA Isolation Kit (Omega Bio-Tek Inc.) and EZNA Total RNA Kit II (Omega Bio-Tek Inc., Norcross, GA, USA), respectively. The expression levels of MET were quantified by quantitative real-time PCR using SYBR Green detection reagent (CoWin Biotech Co., Ltd., Beijing, China) and GAPDH was used as an endogenous control. The relative quantity of mature miR-133b was measured by miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA) according to protocol, U6-snRNA levels were used for normalization. The primers and reaction conditions have been previously described (16).
Western blot analysis
Protein was extracted with complete RIPA buffer, and details about Western blot analysis can be found in our previously studies (16). Briefly, protein samples are boiled and separated on a 10% polyacrylamide gel and transferred to PVDF membranes (Invitrogen, Carlsbad, CA, USA), blocked with 3% BSA for 2 hours and incubated overnight with primary antibody (MET monoclonal antibody, 1:200 dilution; GAPDH monoclonal antibody, 1:200 dilution) (Santa Cruz Biotechnology). After being washed with PBST, immunoreactive bands were visualized through HRP-conjugated m-IgGk secondary antibody, 1:1,000 dilution (Santa Cruz Biotechnology) and ImageQuant LAS 350 (GE Healthcare).
Statistical analysis
Statistical computations were performed using SPSS version 16.0 and GraphPad Prism6. Data coming from experimental are presented as mean ± SD. Paired t-test was used to assess the difference of MET expression between the tumor tissue and non-tumor tissue. The χ2 test was adopted to analyze the association between miR-133b and MET expression and clinicopathological characteristics. Pearson’s correlation analysis was chose for computing the correlation between MET and miR-133b. The diagnostic performance of miR-133b and/or MET was evaluated by receiver operating characteristic (ROC) curve analysis in SPSS version 16.0, and the sensitivity, specificity, and AUC were determined. P>0.05 was considered not statistically significant.
Results
Clinicopathological characteristics of the patients
As summarized in Table 1, the features of clinical and histopathological are presented. There are 46 CRC patients involved in this article (26 males and 20 females, ranging from 55 to 83). Among these patients, out of the 46 patients, 2 (4.35%) patients are well differentiated adenocarcinomas, 28 (60.87%) has moderately differentiated adenocarcinomas, and 16 (34.78%) possess poorly differentiated adenocarcinomas. For the stage of progression of the CRC cases studied, most of the patients were in the advanced stage of the disease: the first stage diagnosis was 6 cases (13.04%), the second stage was 11 cases (23.91%), the third stage was 25 cases (54.35%), the fourth stage was 4 cases (8.69%).
Table 1
| Clinicopathological features | Number | miR-133b | MET | |||||
|---|---|---|---|---|---|---|---|---|
| Lowa (n=23) | Higha (n=23) | P value | Lowa (n=23) | Higha (n=23) | P value | |||
| Age (years) | 46 | |||||||
| ≥60 | 34 | 15 | 19 | 0.397 | 20 | 14 | 0.318 | |
| <60 | 12 | 7 | 5 | 9 | 3 | |||
| Tumor siteb | ||||||||
| Right | 13 | 7 | 6 | 0.528 | 4 | 9 | 0.178 | |
| Left | 7 | 2 | 5 | 3 | 4 | |||
| Rectum | 26 | 13 | 13 | 16 | 10 | |||
| Tumor differentiation | ||||||||
| Well | 2 | 0 | 2 | 0.138 | 1 | 1 | 0.712 | |
| Moderate | 28 | 11 | 17 | 12 | 16 | |||
| Poor | 16 | 10 | 6 | 5 | 11 | |||
| Staging | ||||||||
| I | 6 | 3 | 3 | 0.432 | 4 | 2 | 0.677 | |
| II | 11 | 6 | 5 | 7 | 4 | |||
| III | 25 | 19 | 6 | 12 | 13 | |||
| IV | 4 | 4 | 0 | 0 | 4 | |||
| Lymph node | ||||||||
| N0 | 18 | 5 | 13 | 0.028b | 10 | 8 | 0.032b | |
| N1 | 21 | 8 | 13 | 17 | 4 | |||
| N2 | 7 | 6 | 1 | 2 | 5 | |||
| Distant metastasis | ||||||||
| M0 | 42 | 17 | 25 | 0.022b | 31 | 11 | 0.003b | |
| M1 | 4 | 4 | 0 | 0 | 4 | |||
a, low and high expression groups were determined by the median value of MET and miR-133b mRNA in the tumor tissue specimens; b, statistical significance (P<0.05). CRC, colorectal cancer; MET, mesenchymal-epithelial transition factor.
Downregulation of miR-133b is closely correlated with MET expression in CRC tissues
In order to study the biological functions of miR-133b and MET in human CRC, RT-qPCR was used to analyze the expression of miR-133b and MET mRNA in human CRC tissues and matched healthy counterparts. The expression levels of miR-133b and MET mRNA were compared to these levels in human healthy colon cell line (CCD-18Co cell line serve as a reference) (16). Tumor specimens show decreased miR-133b expression compared to non-tumor specimens (P<0.001, Figure 1A). Conversely, over-activation of MET mRNA in tumor specimens were significantly higher than those in the corresponding non-tumor specimens (P<0.001, Figure 1B).
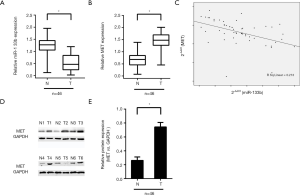
We next expanded our analysis to the relationship between miR-133b expression and MET. A very strong inverse correlation was found between the expression of MET mRNA and the expression of miR-133b (n=46, r=−0.213, P=0.006, Pearson’s correlation; Figure 1C). These experimental data showed an intermodulation of miR-133b and MET, indicating that the downregulation of miR-133b may lead to MET overexpression in CRC tissues.
In addition, western blot analysis was performed to determine whether the protein expression of MET was consistent with the mRNA expression. As demonstrated in the Figure 1D and E, an identical pattern is observed in the expression of MET protein expression.
Association of miR-133b and MET expression with the clinicopathological parameters of CRC tissues
Aberrant expression of MET and miR-133b has been identified in above analysis, which suggesting that there may be a close association between expression of MET and miR-133b with various clinicopathological parameters of the CRC (Table 1). To address this issue, χ2 test was performed and the result found that MET expression was negative associated with lymph node and distant metastases (P<0.05). A similar pattern was observed for MiR-133b expression, its low expression is inversely correlated with lymph node and distant metastases. No statistical significance was found between MET and miR-133b expression and other clinicopathological parameters (P>0.05).
Diagnostic capability of miR-133b and MET expression in CRC
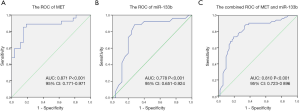
The fact that the aberrant expression of miR-133b and MET presented a close association with clinicopathological parameters of CRC remind us both can be serve as an optimum predictive biomarker for CRC patients. To assess the diagnostic capability of miR-133b and MET expression in CRC, the receiver operator characteristic (ROC) curve was used to further validate the sensitivity and specificity of miR-133b and MET dysregulated expression in indicating the occurrence of CRC. The AUC of MET for CRC was 0.871 [95% confidence interval (CI), 0.771–0.971; P<0.001] (Figure 2A). The AUC of miR-133b for CRC was 0.778 (95% CI, 0.651–0.924, P<0.001) (Figure 2B). The combined AUC of MET and miR-133b for CRC was 0.810 (95% CI, 0.723–0.896, P<0.001) (Figure 2C). These data indicated that MET and miR-133b are suitable predictors for CRC.
Discussion
Elucidating the molecular mechanisms of tumors is beneficial for the treatment of CRC and for many other types of tumors. Multiple sequential steps and many genes are involved in CRC process and there is a complex disorder of gene expression, including both the coding and noncoding gene structure and expression. Over the past decade, a great amount of scientific effort has been put into the discovery of alternative biomarkers that can be employed clinically for early diagnosis and prognosis indicators for CRC patients (20,21). However, to date, none of these potential biomarkers have been prospectively validated and identified. Thus, to significantly improve clinical decision-making, it is evident that there is an urgent need for novel, reliable predictive biomarkers.
MicroRNAs are called ‘oncomirs’ because they function either as tumor suppressors or oncogenes. The investigation of differentially expressed microRNAs in cancer specimens has produced important information on their role in carcinogenesis (22). A series recent of studies have suggested that miR-133b has been identified in various cancers to act as a tumor suppressor gene and has deep relationship to suppressed carcinoma metastasis. The downregulated of miR-133b is confirmed in esophageal squamous cell carcinoma, and miR-133b/EGFR axis regulates cell metastasis by suppressing anoikis resistance and anchorage-independent growth (23). And the miR-133b is downregulated in prostate cancer tissue, via activating the TGF-β signaling, thereby contributing to enhanced invasion and migration abilities in vitro and bone metastasis ability in vivo (24). With the deciphering of roles for miR-133b in this study, we demonstrated a statistically significant downregulation of miR-133b in CRC tissues and the expression of miR-133b is inversely correlated with oncogene MET. Moreover, patients with a low expression of miR-133b and a high expression of MET tend to have advanced clinical stage and have metastasis invasion into lymphatic vessels, which indicate that the expression levels of miR-133b and MET may be useful as predictive biomarkers in CRC.
miR-133b has been frequently reported in many malignant tumors, including gastric, lung, hepatocarcinoma, renal cancer and CRC. Previous studies have focused on cell lines, and the clinical importance of miR-133b in CRC was not characterized until recently (11,12,14,25). In this study, data analysis confirmed that the expression level of miR-133b in CRC tissues was significantly lower than in that of pair-matched adjacent non-tumor tissues; CRC specimens with a relatively lower miR-133b expression tend to exhibit poor cell differentiation and promoted a migratory and invasive phenotype. This evidence suggests that miR-133b may serve as a tumor suppressor gene and that the downregulation of miR-133b expression may have a central regulatory role in the initiation and progression of CRC. Meanwhile, no statistical significance was found with miR-133b expression between cancer tissues and their pair-matched adjacent non-tumor tissues in patient age or gender, tumor site or the extramural vascular invasion status. It is possible that with an increase in sample size different and interesting results may be found.
MET activates many signal transduction pathways including those of RAS, PI3K-Akt, STAT and beta-catenin (19). MET and phosphorylated MET expression is positively correlated with tumor grade, stage, and size and patient survival in multiple types of cancers. It is possible that MET can serve as an optimum therapeutic target in cancer (26-28). Our previous studies have shown that miR-133b inhibits MET expression by targeting the MET 3’UTR in CRC cells (16). This was confirmed by the use of RTqPCR in CRC specimens in this study, which indicated that miR-133b expression was inversely associated with MET expression. The cancer tissues in our studies show increased expression of MET and the expression of it was correlated with advanced tumors, nodes and metastasis stage. Additionally, RT-qPCR and western blot analyses showed that MET was highly expressed in primary tumors from CRC patients. These findings suggest that miR-133b may promote a migratory and invasive phenotype and lead to advanced clinical disease stages by regulating MET in CRC.
MiRNAs can influence the differentiation, growth and apoptosis of normal cells through various pathways, while the abnormal expression of microRNAs can alter the physiological and morphological features of different kinds of cells and induce the abnormal hyperplasia of tissues or tissues to be differentiated into tumors (29,30). Hence, abnormally expressed miRNAs can serve as specific markers for the diagnosis of certain tumors. MicroRNAs have been identified as potential predictive biomarkers in various cancers in different studies (30-32). In this study, the ROC analysis showed that miR-133b and/or MET may be useful for identifying CRC patients from the normal population, indicating that miR-133b and MET may be prospective, non-invasive diagnostic markers for early-stage CRC. However, this result was obtained from CRC tissues, and it remains unclear whether the same results could be obtained in blood samples that can be easily obtained from individuals.
As we demonstrated here, aberrations of miR-133b has been found in CRC tissues and miR-133b can be regarded as a predictive tumor suppressor for CRC. The conclusion of low expression of miR-133b and high expression of MET being positively correlated with advanced clinicopathological features such as migratory and invasive phenotype suggest that miR-133b and MET may serve as potential valuable and optimum diagnostic biomarkers for CRC.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2370
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2370
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2370). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Central South University (approval number: No. 2016-s104). The patients gave informed consent before taking part.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet 2016;7:105-14. [PubMed]
- Li W, Qiu T, Ling Y, et al. Molecular pathological epidemiology of colorectal cancer in Chinese patients with KRAS and BRAF mutations. Oncotarget 2015;6:39607-13. [Crossref] [PubMed]
- Tokodai K, Narimatsu H, Nishida A, et al. Risk factors for recurrence in stage II/III colorectal cancer patients treated with curative surgery: The impact of postoperative tumor markers and an infiltrative growth pattern. J Surg Oncol 2016;114:368-74. [Crossref] [PubMed]
- Julien M, Dove J, Quindlen K, et al. Evolution of Laparoscopic Surgery for Colorectal Cancer: The Impact of the Clinical Outcomes of Surgical Therapy Group Trial. Am Surg 2016;82:685-91. [Crossref] [PubMed]
- Bradatsch A, Kornprat P, Bacher H, et al. The Value of Lymph Node Dissection in the Surgery of Colorectal Cancer Liver Metastases. Anticancer Res 2016;36:2993-7. [PubMed]
- Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 2005;11:1753-61. [Crossref] [PubMed]
- Slack FJ, Weidhaas JB. MicroRNA in cancer prognosis. N Engl J Med 2008;359:2720-2. [Crossref] [PubMed]
- Pal MK, Jaiswar SP, Dwivedi VN, et al. MicroRNA: a new and promising potential biomarker for diagnosis and prognosis of ovarian cancer. Cancer Biol Med 2015;12:328-41. [PubMed]
- Wang B, Shen ZL, Jiang KW, et al. MicroRNA-217 functions as a prognosis predictor and inhibits colorectal cancer cell proliferation and invasion via an AEG-1 dependent mechanism. BMC Cancer 2015;15:437. [Crossref] [PubMed]
- Cao ZG, Li JJ, Yao L, et al. High expression of microRNA-454 is associated with poor prognosis in triple-negative breast cancer. Oncotarget 2016;7:64900-9. [Crossref] [PubMed]
- Li H, Xiang Z, Liu Y, et al. MicroRNA-133b Inhibits Proliferation, Cellular Migration, and Invasion via Targeting LASP1 in Hepatocarcinoma Cells. Oncol Res 2017;25:1269-82. [Crossref] [PubMed]
- Yang X, Lei P, Huang Y, et al. MicroRNA-133b inhibits the migration and invasion of non-small cell lung cancer cells via targeting FSCN1. Oncol Lett 2016;12:3619-25. [Crossref] [PubMed]
- Chang L, Lei X, Qin YU, et al. MicroRNA-133b inhibits cell migration and invasion by targeting matrix metalloproteinase 14 in glioblastoma. Oncol Lett 2015;10:2781-6. [Crossref] [PubMed]
- Guo L, Bai H, Zou D, et al. The role of microRNA-133b and its target gene FSCN1 in gastric cancer. J Exp Clin Cancer Res 2014;33:99. [Crossref] [PubMed]
- Wu D, Pan H, Zhou Y, et al. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol Med Rep 2014;9:2491-8. [Crossref] [PubMed]
- Hu G, Chen D, Li X, et al. miR-133b regulates the MET proto-oncogene and inhibits the growth of colorectal cancer cells in vitro and in vivo. Cancer Biol Ther 2010;10:190-7. [Crossref] [PubMed]
- Gherardi E, Sandin S, Petoukhov MV, et al. Structural basis of hepatocyte growth factor/scatter factor and MET signalling. Proc Natl Acad Sci U S A 2006;103:4046-51. [Crossref] [PubMed]
- Tolbert WD, Daugherty-Holtrop J, Gherardi E, et al. Structural basis for agonism and antagonism of hepatocyte growth factor. Proc Natl Acad Sci U S A 2010;107:13264-9. [Crossref] [PubMed]
- Matsumoto K, Umitsu M, De Silva DM, et al. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci 2017;108:296-307. [Crossref] [PubMed]
- Wada H, Shiozawa M, Katayama K, et al. Systematic review and meta-analysis of histopathological predictive factors for lymph node metastasis in T1 colorectal cancer. J Gastroenterol 2015;50:727-34. [Crossref] [PubMed]
- Zeng W, Tu Y, Zhu Y, et al. Predictive power of circulating miRNAs in detecting colorectal cancer. Tumour Biol 2015;36:2559-67. [Crossref] [PubMed]
- Ji W, Sun B, Su C. Targeting MicroRNAs in Cancer Gene Therapy. Genes (Basel) 2017;8:21. [Crossref] [PubMed]
- Zhu JF, Liu Y, Huang H, et al. MicroRNA-133b/EGFR axis regulates esophageal squamous cell carcinoma metastases by suppressing anoikis resistance and anchorage-independent growth. Cancer Cell Int 2018;18:193. [Crossref] [PubMed]
- Huang S, Wa Q, Pan J, et al. Transcriptional downregulation of miR-133b by REST promotes prostate cancer metastasis to bone via activating TGF-β signaling. Cell Death Dis 2018;9:779. [Crossref] [PubMed]
- Li D, Xia L, Chen M, et al. miR-133b, a particular member of myomiRs, coming into playing its unique pathological role in human cancer. Oncotarget 2017;8:50193-208. [Crossref] [PubMed]
- Ma PC. Special issue: "MET as actionable target in cancer therapy Ann Transl Med 2017;5:1. [Crossref] [PubMed]
- Shali H, Ahmadi M, Kafil HS, et al. IGF1R and c-met as therapeutic targets for colorectal cancer. Biomed Pharmacother 2016;82:528-36. [Crossref] [PubMed]
- Rothenberger NJ, Stabile LP. Hepatocyte Growth Factor/c-Met Signaling in Head and Neck Cancer and Implications for Treatment. Cancers (Basel) 2017;9:39. [Crossref] [PubMed]
- Orellana EA, Kasinski AL. MicroRNAs in Cancer: A Historical Perspective on the Path from Discovery to Therapy. Cancers (Basel) 2015;7:1388-405. [Crossref] [PubMed]
- Jiang C, Chen X, Alattar M, et al. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther 2015;22:291-301. [Crossref] [PubMed]
- Tessitore A, Cicciarelli G, Mastroiaco V, et al. Therapeutic Use of MicroRNAs in Cancer. Anticancer Agents Med Chem 2016;16:7-19. [Crossref] [PubMed]
- Xuan Y, Yang H, Zhao L, et al. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett 2015;360:89-105. [Crossref] [PubMed]


