
Identification of six hub genes and analysis of their correlation with drug sensitivity in acute myeloid leukemia through bioinformatics
Introduction
Acute myeloid leukemia (AML) is the most malignant type of leukemia, with new cases increasing every year worldwide and as well as with age (1), which is a class of hematopoietic stem cells, malignant clonal diseases derived from progenitor cells. Abnormal primitive cells and partial primitive immature cells (leukemia cells) in bone marrow (BM) proliferate in large quantities and inhibit normal hematopoiesis, resulting in BM failure and rapid progress (2).
The prognosis of current AML patients is not optimistic, and only 40% of patients under the age of 60 survive more than 5 years. Although most patients can achieve remission after initial chemotherapy, patients who relapse after complete response rarely survive more than 5 years (3,4). Chemotherapy and hematopoietic stem cell transplantation (HSCT) have been the most important treatment for AML in recent decades (5,6). Nevertheless, the drug resistance is increasingly hindering the therapeutic effect of AML patients, so that the quality of life of these patients is relatively poor. At the same time, recurrence remains the most common cause of HSCT failure in patients with AML, as a result, the prognosis for these patients remains poor (5). Moreover, there is increasing evidence that abnormal gene expression and mutated genes are involved in the carcinogenesis, progression and prognosis of AML. Therefore, there is an urgent need to find AML biomarkers and develop sensitive therapeutic drugs.
Over the past several decades, with the rapid rising of precision medicine, chip technology and bioinformatics analysis have been widely used to screen genetic changes at the genome level, which help us identify the differentially expressed genes (DEGs) associated with the carcinogenesis and progression of AML, and then find the precise sensitive chemotherapy drugs and improve patient outcomes. Consequently, in this study, in order to obtain reliable results, we downloaded two mRNA chip datasets (GSE9476 and GSE30029) from Gene Expression Omnibus (GEO) for analysis, and obtained the DEGs between AML patients and normal BM tissues. After a series of screening and validation, 456 DEGs were identified and six hub genes (FLT3, PF4, CD163, MRC1, CSF2RB, PPBP) were obtained. And then, the relationship between drug sensitivity and gene expression of the six hub genes was analyzed in AML cell lines. The study provides preliminary evidences for search the potential sensitive therapeutic drugs for AML.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2712).
Methods
Data processing for microarray data and DEGs
Two separate gene expression profiles [GSE9476 (7) and GSE30029 (8,9)] were chosen from GEO database (http://www.ncbi.nlm.nih.gov/geo/), and GSE9476 was based on the GPL96 platform, and GSE30029 was based on the GPL6947 platform. Then, the DEGs through GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) online analysis tool in NCBI were screened (10), and identified with fold change|FC| ≥1, and defined P value and false discovery rate (FDR) cutoffs of <0.05. Then, the Venn diagram was made through the web tool Bioinformatics & Systems Biology (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Protein-protein interaction (PPI) network construction and hub gene identification
The PPI map was obtained by the Retrieval of Interacting Genes (STRING) database (https://string-db.org/), an online tool allowing users uploading the data of DEGs. It can be used to analyze the PPI information and to evaluate the interactive relationships among DEGs (11). The PPI pairs were set the combined score >0.4. These pairs were extracted and visualized for the PPI network using Cytoscape (3.7.2) software (www.cytoscape.org/). In Cytoscape, module screening was performed using the Molecular Complex Detection (MCODE) (scores >3 and nodes >4), and the degree of connection was computed using the Centralities for Cytoscape (Centiscape2.2) (12). Nodes with higher degree of connection were more essential for maintaining the stability of the entire network, usually, nodes with degree of connection ≥10 were considered to be core candidate genes.
GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis
To evaluate the functional annotation and enrichment analysis of DEGs, the Database for Annotation, Visualization and Integrated Discovery (DAVID) tools (https://david.ncifcrf.gov/) were used (13,14). From the DAVID v6.8 online server, the functional annotation and enrichment analysis was performed by the Gene Ontology (GO). The biological process, molecular function, and cellular component are the three major components of gene function. KEGG collects large number of data about molecular-level information, biological pathways, chemical substances through high-throughput experimental technologies. The KEGG pathway was analyzed from the DAVID. P<0.05 and gene counts ≥10 were considered statistically significant.
Validation and screening of key genes using GEPIA2
The Gene Expression Profiling Interactive Analysis (GEPIA) is an online database of gene expression analysis, which based on tumor and normal samples from The Cancer Genome Atlas (TCGA; http://portal.gdc.cancer.gov/) and the Genotype-Tissue Expression (GTEx) (https://www.gtexportal.org/) projects. GEPIA2 is an updated and enhanced version, which extends gene expression quantification from the gene level to the transcript level and supports analysis of a specific cancer subtype and comparison between subtypes (15). The expression of hub genes between AML and normal BM tissues was verified by the database. Expression DIY (box plot) and survival analysis were performed. The parameters of the expression DIY (box plot) were set as follows: |log2FC| cutoff =1, P value cutoff =0.01, log scale = yes, jitter size =0.4, match TCGA normal and GTEx data. The parameters of the survival analysis were set as follows: overall survival (OS), group cutoff median, cutoff-high (%) =50, cutoff-low (%) =50, hazards ratio (HR) = yes, 95% confidence interval = yes, axis units months. According to the following screening criteria: the expression DIY (box plot) indicated that the gene was higher expressed in the AML sample than normal BM and peripheral monocytes, the survival analysis (OS) of the gene indicated a lower prognosis and must be statistically significant (P<0.05). The BloodSpot database (16) (http://servers.binf.ku.dk/bloodspot/) is a public microarray database for mRNA expression of BM and gene-centric blood cells. The correlation between the expression of key genes and the genetic characteristics of AML patients was analyzed using the BloodSpot database.
The relationship between genes expression and drug sensitivity in GSCA Lite
GSCA Lite (http://bioinfo.life.hust.edu.cn/web/GSCALite/) is a web-based platform for Gene Set Cancer Analysis, which is dynamic analysis and visualization of gene set in expression of malignant tumor genes and drug sensitivity correlation. The Drug Sensitivity Analysis of GSCA Lite has collected 481 small molecules from Cancer Therapeutics Response Portal (CTRP) (https://portals.broadinstitute.org/ctrp/) and 265 small molecules from Genomics of Drug Sensitivity in Cancer (GDSC) (https://www.cancerrxgene.org/) (17,18). Drug sensitivity and gene expression profiling data of cancer cell lines in GDSC and CTRP are integrated for investigation (19). The expression of each gene in the gene set is performed by Spearman correlation analysis with the small molecule/drug sensitivity (IC50). The Spearman correlation represents the gene expression correlates with the drug. The positive correlation means that the gene high expression is resistant to the drug, vise versa.
Statistical analysis
The results of this study were downloaded from the database according to the optimal statistical method selected by the database itself. Original chip data were downloaded from GEO databases for statistical analysis using unpaired sample t-test. The results were expressed as mean ± labeled standard deviation, and P<0.05 indicated that the results were statistically significant.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Identification of DEGs based on clinical samples of GEO in AML
In our study, we selected two datasets: GSE9476 and GSE30029. GSE9476 contained 26 samples of AML patients and 20 samples of healthy volunteers, while GSE30029 contained 90 samples of AML patients and 31 samples of healthy volunteers. According to the criteria of P<0.05 and |log FC| ≥1, 2,058 DEGs were screened out in GSE9476 and 3653 DEGs were screened out in GSE30029 through comparing AML patient samples with healthy volunteers. We made a Venn diagram to show the association between the DEGs of GSE9476 and GSE30029. The 456 DEGs were the overlaps of the two datasets, as shown in Figure 1A.
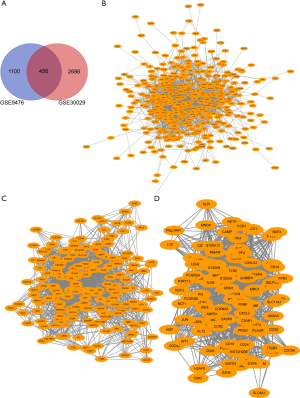
KEGG and GO enrichment analysis, PPI network and key gene identification of DEGs
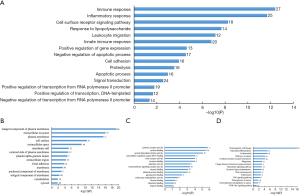
Among 456 DEGs, the PPI action network was shown in Figure 1B. Commonly, only connectivity degree ≥10 can be identified as potential core gene, accordingly, among 456 DEGs, 183 potential core genes which have been calculated by using Cytoscape (3.7.2) software met the condition of degree of connection ≥10 (Figure 1C). Then, the possible association among the183 DEGs was evaluated by STRING online tool. PPI network was shown in Figure 1C. Meanwhile, the top 100 genes of the degree of connection of these 183 genes were selected and shown in Figure 1D (Table S1). We found that the degree of tumor necrosis factor (TNF) was 106, which ranked first among these 100 genes. Then, the following genes were integrin subunit alpha M (ITGAM; degree =101), protein tyrosine phosphatase, receptor type C (PTPRC; degree =83), toll like receptor 2 (TLR2; degree =74), matrix metallopeptidase 9 (MMP9; degree =74) and so on. These 100 genes could be the core genes screened out in AML. The enrichment analysis of gene function and pathway of these 183 DEGs were carried out by DAVID v6.8 online server, and the GO analysis and KEGG pathway analysis results were shown in (Figure 2). The most significant enrichment for DEGs in biological processes was in immune response with the enrichment count of 27 genes, which was the highest number of gene DEGs. The second is in inflammatory response (Figure 2A). The most significant enrichment for DEGs in cell component was in integral component of plasma membrane with the enrichment count of 69. The second was in extracellular exosome with gene enrichment number of 75 (Figure 2B). The most significant differentially expressed enrichment in molecular function was cytokine receptor activity, and the count of gene enrichment was 7. The second was protein binding, and the count of gene enrichment was 123, which was the count of gene enrichment, and so on (Figure 2C). KEGG pathway analysis revealed that the most significant enrichment for DEGs was related to hematopoietic cell lineage, in which there was the enrichment count of 21 (Figure 2D).
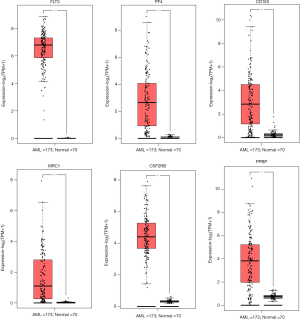
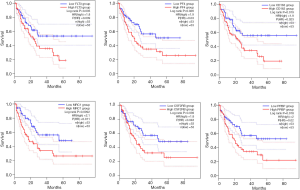
The relationship between hub genes and prognosis of AML patients by GEPIA2
To further verify, screen and analyze the expression of the top 100 genes of DEGs in peripheral blood mononuclear and BM cells of AML patients and healthy volunteers, the expression DIY (box plot) and the survival analysis by GEPIA2 were performed. Accordingly, as shown in Figure 3, six higher expressed genes (FLT3, PF4, CD163, MRC1, CSF2RB, PPBP) were found in the AML compared with healthy volunteers (P<0.05). At the same time, as in Figure 4, the survival analysis (OS) indicated there was reverse relationship between the expression level of the above six genes and OS (P<0.05). The result suggests the six highly expressed genes might predict a worse prognosis in AML patients (P<0.05).
FLT3, PF4, CD163, MRC1, CSF2RB and PPBP may be the potential hub genes of AML
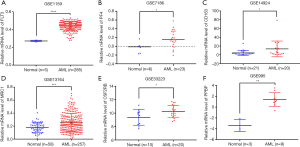
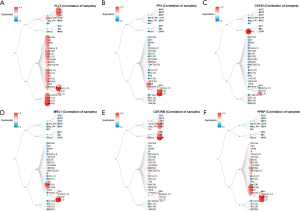
To further verify the differentially expression of the selected six candidate genes between AML patients and normal BM tissues, mRNA expression of FLT3, PF4, CD163, MRC1, CSF2RB and PPBP at mRNA levels comparing with AML samples and healthy volunteers of peripheral blood and/or BM mononuclear cells was performed through other microarray datasets in GEO database (Figure 5). We validated that they were higher expressed in AML samples obviously than control, which were statistically significant (P<0.05). In addition, as the recommended genetic classification of AML includes low-risk group, intermediate-risk group and high-risk group (20). In addition, the BloodSpot database was used to analyze the effect of hub genes expression and genetic abnormalities on risk stratification in AML patients (Figure 6). We found that high expression of these six hub genes in AML patients may indicate high-risk groups at the genetic stratification level.
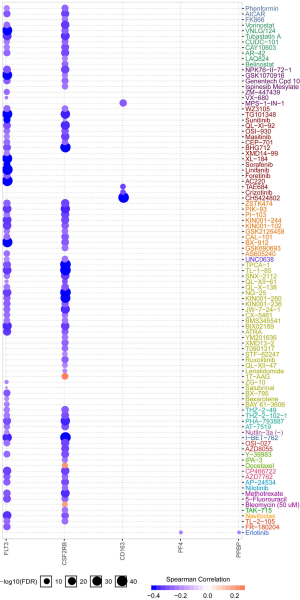
The correlation of hub genes expression and drug sensitivity in AML cells by GSCA Lite
To analyze the correlation of six hub genes (FLT3, PF4, CD163, MRC1, CSF2RB, PPBP) expression in AML and the sensitivity of AML cells to the small-molecule drugs, we used the GDSC IC50 drug data from GSCA Lite database (Figure 7). We found that AML cell lines with FLT3-overexpression and CSF2RB-overexpression were sensitive to most small-molecule drugs which can also be used in malignancies including target drugs and non-target drugs, while AML cell lines with CD163-overexpression were only sensitive to CH5424802, Crizotinib, TAE684 and MPS1-IN-1. And Erlotinib (Tarceva) was sensitive for AML cell lines with PF4-overexpression and PPBP-overexpression. For example, looking up from the GDSC database, we discovery that Sorafenib (Nexavar), whose targets are PDGFR, KIT, VEGFR, RAF and FLT3. And in our study, we verified that its effect on AML cell lines with FLT3-overexpression was sensitive, and AML cell lines with high FLT3 expression were also sensitive to the other small molecule drugs such as all-trans retinoic acid (ATRA). In our analysis, AML cells with the overexpression PF4 and PPBP displayed sensitivity to Erlotinib (Tarceva), whose drug target is EGFR from GDSC database. The sensitivity to methotrexate, which is an antimetabolite drug, is correlated with the high expression of CSF2RB in AML cell lines. Details of common small-molecule drug were shown in Table S2.
Discussion
AML is one of the most common and aggressive type of leukemia, and the treatment of AML is given priority to combination chemotherapy. The treatment of AML also has made progress in last decade, however, recurrence rates remained still high (3,4,6). So, searching for accurate molecular biomarkers is urgently needed. Microarray technology has enabled us to explore genetic changes in diseases and has been shown to be an effective way to identify novel biomarkers for human tumor including AML.
In the present study, 2 mRNA microarray datasets were analyzed to obtain DEGs between AML and healthy subjects of peripheral blood and BM samples. A total of 456 DEGs were identified among the 2 datasets. Then, we performed DAVID enrichment analysis on these 456 DEGs, GO and KEGG enrichment analyses were performed to explore interactions among the 456 DEGs. GO enrichment analysis revealed that changes in the most significant modules were mainly enriched in regulation of transcription from RNA polymerase II promoter, DNA-templated, cell proliferation and division. While, changes in KEGG were mainly enriched in viral carcinogenesis, transcriptional mis-regulation in cancer, cell cycle and p53 signaling pathway. Previous studies have reported that the dysregulation of transcription from RNA polymerase II promoter, cell proliferation, cell division and immune response play important roles in the carcinogenesis or progression of tumors (21-23). Similarly, transcriptional mis-regulation in cancer, cell cycle and p53 signaling pathway have been shown in previous studies to play a very important role in the occurrence and development of tumors (22,24). Therefore, our results suggest that the DEGs played an important role in AML tumorigenesis and progression, which is consistent with the previous studies.
Through a series of screening, analysis and verification, we screened 6 DEGs (FLT3, PF4, CD163, MRC1, CSF2RB, PPBP) as the hub genes. In BM and peripheral blood samples from AML patients, the expression of the hub genes was higher than healthy volunteers. In terms of the relationship between the expression of individual hub genes and patient prognosis, the OS rate of AML patients with higher expression of the above six hub genes was lower. Therefore, not only FLT3, PF4, CD163, MRC1, CSF2RB and PPBP may be AML oncogenes but also AML patients with higher expression of these six hub genes might have poor prognosis, as well as they might be potential biomarkers of AML. In addition, high expression of these six hub genes in AML patients may indicate high-risk groups at the genetic stratification level, which will provide a theoretical basis for the treatment of frontline doctors.
We found that AML cells with high expression of FLT3 and CSF2RB were sensitive to most small molecule drugs by bioinformatics analysis. While, AML patients with high expression of CD163 were only sensitive to a few small molecule drugs. Only was Erlotinib (Tarceva) related with sensitive in AML cell lines with high PF4 and PPBP expression. With traditional chemotherapy drugs, adaptive resistance existed in many AML patients, leading to poor prognosis for AML patients. There was a median survival of only 4 months for relapsed and/or refractory AML (25,26). So, it was urgent to find novel biomarkers and new therapeutic sensitive drugs for high-risk AML. In our study, the panel included FLT3, PF4, CD163, MRC1, CSF2RB and PPBP might be the novel biomarkers for target therapy in AML.
FLT3, located in 13q12.2, which is an important cytokine receptor, encodes a class III receptor tyrosine kinase and mainly expresses in the hematopoietic compartment and involved in the apoptosis, proliferation and differentiation of hematopoietic cells (26,27). FLT3 mutations which occur in 30% of AML patients, may lead to the occurrence and development of acute leukemia, especially AML (28). In our study, the analysis from microarray datasets showed FLT3 was upregulated in AML and the AML patients with the high expression of the FLT3 gene had a worse prognosis. The result is in agreement with previous reports that high transcript levels of FLT3 were associated with a worse prognosis in AML and pediatric AML patients (29,30). FLT3 inhibitors (tyrosine kinase inhibitors) have been considered to be a target for anti-cancer drugs, which have been used in preclinical and clinical studies, such as sorafenib, lestaurtinib and sunitinib, and so on (26). We analyzed the association between the drug sensitivity of AML cells and the gene expression of these six hub genes using the GSCA Lite database. Our current results showed that besides being sensitive to drugs targeting FLT3, AML patients with high FLT3 expression were also sensitive to the other small molecule drugs such as ATRA. The studies of Ma et al. revealed combining ATRA and FLT3 TKIs had highly synergistic drug activity to eliminate FLT3/internal tandem duplication (ITD)+ leukemia stem cells in vitro and in vivo (31). PF4 (platelet factor 4, CXCL4, located in 4q13.3), encoding the protein of members of the CXC chemokine family. This protein is chemotactic for numerous other cell type and also functions as an inhibitor of hematopoiesis, angiogenesis and T-cell function. It is involved in regulating the differentiation of hematopoietic stem cells/progenitor cells, maintaining the survival and self-renewal of hematopoietic stem cells, and regulating the cardiac differentiation potential of induced pluripotent stem cells (32,33). Previous studies have shown that PF4 is an endocrine factor, and its overexpression in tumors is associated with decreased OS of patients (34). The analysis of Chen et al. from the transcriptome database showed that the high expression of PF4 was a risk factor for poor prognosis of patients (35). However, the study of Bai et al. indicated that PF4 protein was significantly decreased in in newly diagnosed AML group, compared with healthy control group (36). By database analysis, our results showed that the PF4 transcription level of AML patients was higher than that of healthy controls and the OS rates of AML patients with high PF4 expression were shorter compared with low expression of PF4. But further study is required to characterize the function of the gene involved in AML (34,35,37). CD163 (the macrophage scavenger receptor, located in 13q12.2) encodes a protein that clears the scavenger receptor cysteine-rich (SRCR) superfamily. It functions as an acute phase-regulated receptor involved in the clearance and endocytosis of hemoglobin/haptoglobin complexes by macrophages. Its overexpression is associated with associated with a poor prognosis in AML and the immune/inflammatory response, which is potential biomarker of myeloid leukemia cutis (38,39). At the same time, CD163 is also involved in the development of multiple myeloma, meningioma, Hodgkin lymphoma and colorectal cancer (40-43). Additionally, CD163 is regarded as a potential therapeutic target for cell-directed therapy on macrophages in some cancers, such as glioma, gastric cancer (44-46). In our study, CD163 was identified to be significantly associated with AML prognosis by integrated bioinformatics analysis, which was consistent with the results of previous studies and would provide a preliminary basis for further development of targeted drugs for AML with high expression of CD163. MRC1 (mannose receptor C-type 1, CD206, located in 10p12.33), a member of the C-type lectin receptor family, encodes the human mannose receptor (MR). It is involved in several biological processes, including cell-cell recognition, serum glycoprotein turnover, and neutralization of pathogens. MRC1 mutations can cause activation and replication defects at DNA damage checkpoints, which are associated with tumorigenesis (47). MRC1 can also regulate tumor microenvironment (48), the relationship between tumor recurrence and metastasis (49). In our study, we have preliminarily confirmed that MRC1 is higher expressed in AML compared to healthy volunteers and is a potential biomarker for AML, which laid a solid foundation for our further development of targeted drugs targeting MRC1 overexpression in AML. CSF2RB (colony-stimulating factor 2 receptor subunit beta, CD131) is located in 22q12.3. The protein encoded by CSF2RB is the common beta chain of the high affinity receptor for IL-3, IL-5 and CSF. It is a myeloid cytokine receptor, which is highly expressed in myeloid leukemia of Down syndrome (ML-DS), and CSF2RB variant A455D is an oncogenic hotspot mutation in ML-DS (50). The high expression of CSF2RB predicts the arrest of hematopoietic stem cell differentiation and CSF2RB was expressed at high levels in both mouse and human leukemia cells (51). In our study, we found CSF2RB was over-expressed and predicted worse prognosis in AML. PPBP (pro-platelet basic protein, CXCL7, located in 10p12.33) is a platelet-derived growth factor that belongs to the CXC chemokine family. It has been shown to stimulate various cellular processes including DNA synthesis, mitosis, glycolysis, intracellular cAMP accumulation, prostaglandin E2 secretion, and synthesis of hyaluronic acid and sulfated glycosaminoglycan. Previous studies have implicated serum levels of CXCL7 as potential prognostic markers in myelodysplastic syndromes and the overexpression of the gene in the marrow microenvironment contributed to induction of decitabine resistance in chronic myelomonocytic leukemia cells (52,53). In addition, PPBP is a biomarker in gastric cancer (54) and a predictive marker of sunitinib efficacy in clear cell renal cell carcinomas (55). The results of our analysis indicated AML patients with the high expression of PPBP gene had worse prognosis and AML cells with the overexpression PPBP displayed sensitivity to Erlotinib (Tarceva). Therefore, in present study, we thought CSF2RB and PPBP might also be a potential biomarker in AML.
Most of the six hub genes and their correlation with the sensitive drug we have derived from the study have been confirmed by previous studies to be associated with AML or cancer. But there are still several hub genes and their correlation with the sensitive drug have not been explored to provide a basis for guiding clinical medication, which may be the innovation of future research. Our study may not only provide potentially likely regulators of AML invasion and metastasis can serve as biomarkers in AML, also can give future researchers a broader perspective and more inspiration. But it still has limitation: (I) the analysis of drug sensitivity was based on the analysis of AML cell lines, which needs to be further verified in animal experiments; (II) the analysis of DEGs used in this study is based on the comprehensive analysis of clinical case samples, which needs to be further verified in animal experiments and other preclinical trial. Next, we will further verify it through animal experiments and other preclinical trial, which will be a significant and constructive study. Also, we will conduct further in-depth research on the expression and mechanism of these six hub genes in AML.
Conclusions
Our study was designed to identify DEGs that might be involved in the carcinogenesis or progression of AML. A total of 456 DEGs and six hub genes (FLT3, PF4, CD163, MRC1, CSF2RB and PPBP) were identified and might be regarded as promising biomarkers and potential sensitive drugs were identified for overexpression of the biomarkers in AML, and high expression of these six hub genes in AML patients may indicate high-risk groups at the genetic stratification level, which will provide a theoretical basis for the treatment of frontline doctors.
Acknowledgments
Funding: Open Access funding provided by
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2712
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2712
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2712). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The figures and data downloaded for this study were approved by the relevant databases.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rao AV. Fitness in the elderly: how to make decisions regarding acute myeloid leukemia induction. Hematology Am Soc Hematol Educ Program 2016;2016:339-47. [Crossref] [PubMed]
- Estey EH. Acute myeloid leukemia: 2014 update on risk-stratification and management. Am J Hematol 2014;89:1063-81. [Crossref] [PubMed]
- Burnett A, Wetzler M, Löwenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol 2011;29:487-94. [Crossref] [PubMed]
- Stein EM, Tallman MS. Emerging therapeutic drugs for AML. Blood 2016;127:71-8. [Crossref] [PubMed]
- Dillon R, Hills RK, Freeman SD, et al. Molecular MRD status and outcome after transplantation in NPM1 mutated AML. Blood 2020;135:680-8. [Crossref] [PubMed]
- Roboz GJ. Current treatment of acute myeloid leukemia. Curr Opin Oncol 2012;24:711-9. [Crossref] [PubMed]
- Stirewalt DL, Meshinchi S, Kopecky KJ, et al. Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer 2008;47:8-20. [Crossref] [PubMed]
- de Jonge HJ, Woolthuis C, Vos A, et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011;25:1825-33. [Crossref] [PubMed]
- Bosman MCJ, Schepers H, Jaques J, et al. The TAK1-NF-κB axis as therapeutic target for AML. Blood 2014;124:3130-40. [Crossref] [PubMed]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 2012;40:4288-97. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-D452. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Dennis G Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4:3. [Crossref] [PubMed]
- Sherman BT. DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 2007;8:426. [Crossref] [PubMed]
- Tang Z, Kang B, Li C, et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 2019;47:W556-60. [Crossref] [PubMed]
- Bagger FO, Kinalis S, Rapin N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res 2019;47:D881-5. [Crossref] [PubMed]
- Liu CJ, Hu FF, Xia MX, et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 2018;34:3771-2. [Crossref] [PubMed]
- Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 2016;12:109-16. [Crossref] [PubMed]
- Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483:570-5. [Crossref] [PubMed]
- Noort S, Zimmermann M, Reinhardt D, et al. Prognostic impact of t(16;21)(p11;q22) and t(16;21)(q24;q22) in pediatric AML: a retrospective study by the I-BFM Study Group. Blood 2018;132:1584-92. [Crossref] [PubMed]
- Alsuliman A, Muftuoglu M, Khoder A, et al. A subset of virus-specific CD161(+) T cells selectively express the multidrug transporter MDR1 and are resistant to chemotherapy in AML. Blood 2017;129:740-58. [Crossref] [PubMed]
- Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell 2013;152:1237-51. [Crossref] [PubMed]
- Rishi L, Hannon M, Salome M, et al. Regulation of TRIB2 by an E2F1-C/EBPα feedback loop in AML cell proliferation. Blood 2014;123:2389-400. [Crossref] [PubMed]
- Krejci O, Wunderlich M, Geiger H, et al. P53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood 2008;111:2190-9. [Crossref] [PubMed]
- Melgar K, Walker MM, Jones LM, et al. Overcoming adaptive therapy resistance in AML by targeting immune response pathways. Sci Transl Med 2019;11:eaaw8828. [Crossref] [PubMed]
- Wu M, Li C, Zhu X. FLT3 inhibitors in acute myeloid leukemia. J Hematol Oncol 2018;11:133. [Crossref] [PubMed]
- Kazi JU, Rönnstrand L. FMS-like tyrosine kinase 3/FLT3: from basic science to clinical implications. Physiol Rev 2019;99:1433-66. [Crossref] [PubMed]
- Mizuki M, Fenski R, Halfter H, et al. FLT3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood 2000;96:3907-14. [Crossref] [PubMed]
- Kang HJ, Lee JW, Kho SH, et al. High transcript level of FLT3 associated with high risk of relapse in pediatric acute myeloid leukemia. J Korean Med Sci 2010;25:841-5. [Crossref] [PubMed]
- Kuchenbauer F, Kern W, Schoch C, et al. Detailed analysis of FLT3 expression levels in acute myeloid leukemia. Haematologica 2005;90:1617-25. [PubMed]
- Ma HS, Greenblatt SM, Shirley CM, et al. All-trans retinoic acid synergizes with FLT3 inhibition to eliminate FLT3/ITD+ leukemia stem cells in vitro and in vivo. Blood 2016;127:2867-78. [Crossref] [PubMed]
- Ohashi F, Miyagawa S, Yasuda S, et al. CXCL4/PF4 is a predictive biomarker of cardiac differentiation potential of human induced pluripotent stem cells. Sci Rep 2019;9:4638. [Crossref] [PubMed]
- Sinclair A, Park L, Shah M, et al. CXCR2 and CXCL4 regulate survival and self-renewal of hematopoietic stem/progenitor cells. Blood 2016;128:371-83. [Crossref] [PubMed]
- Pucci F, Rickelt S, Newton AP, et al. PF4 promotes platelet production and lung cancer growth. Cell Rep 2016;17:1764-72. [Crossref] [PubMed]
- Chen XM, Zhang HM, Yang B, et al. Analysis of unfavorable prognosis gene markers in patients with acute myeloid leukemia by multiomics. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2019;27:331-8. [PubMed]
- Bai J, He A, Zhang W, et al. Potential biomarkers for adult acute myeloid leukemia minimal residual disease assessment searched by serum peptidome profiling. Proteome Sci 2013;11:39. [Crossref] [PubMed]
- Ruytinx P, Proost P, Struyf S. CXCL4 and CXCL4L1 in cancer. Cytokine 2018;109:65-71. [Crossref] [PubMed]
- Harms PW, Bandarchi B, Ma L. CD163 expression in leukemia cutis. J Cutan Pathol 2010;37:953-7. [Crossref] [PubMed]
- Yan H, Qu J, Cao W, et al. Identification of prognostic genes in the acute myeloid leukemia immune microenvironment based on TCGA data analysis. Cancer Immunol Immunother 2019;68:1971-8. [Crossref] [PubMed]
- Hsi ED, Li H, Nixon AB, et al. Serum levels of TARC, MDC, IL-10, and soluble CD163 in Hodgkin lymphoma: a SWOG S0816 correlative study. Blood 2019;133:1762-5. [Crossref] [PubMed]
- Kanno H, Nishihara H, Wang L, et al. Expression of CD163 prevents apoptosis through the production of granulocyte colony-stimulating factor in meningioma. Neuro Oncol 2013;15:853-64. [Crossref] [PubMed]
- Kvorning SL, Nielsen MC, Andersen NF, et al. Circulating extracellular vesicle-associated CD163 and CD206 in multiple myeloma. Eur J Haematol 2020;104:409-19. [Crossref] [PubMed]
- Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer 2019;18:64. [Crossref] [PubMed]
- Chen T, Chen J, Zhu Y, et al. CD163, a novel therapeutic target, regulates the proliferation and stemness of glioma cells via casein kinase 2. Oncogene 2019;38:1183-99. [Crossref] [PubMed]
- Cheng Z, Zhang D, Gong B, et al. CD163 as a novel target gene of STAT3 is a potential therapeutic target for gastric cancer. Oncotarget 2017;8:87244-62. [Crossref] [PubMed]
- Van Gorp H, Delputte PL, Nauwynck HJ. Scavenger receptor CD163, a Jack-of-all-trades and potential target for cell-directed therapy. Mol Immunol 2010;47:1650-60. [Crossref] [PubMed]
- Srivatsan A, Li BZ, Szakal B, et al. The SWR1 chromatin-remodeling complex prevents genome instability induced by replication fork progression defects. Nat Commun 2018;9:3680. [Crossref] [PubMed]
- Enninga EAL, Chatzopoulos K, Butterfield JT, et al. CD206-positive myeloid cells bind galectin-9 and promote a tumor-supportive microenvironment. J Pathol 2018;245:468-77. [Crossref] [PubMed]
- Zhang C, Yu X, Gao L, et al. Noninvasive imaging of CD206-positive M2 macrophages as an early biomarker for post-chemotherapy tumor relapse and lymph node metastasis. Theranostics 2017;7:4276-88. [Crossref] [PubMed]
- Labuhn M, Perkins K, Matzk S, et al. Mechanisms of progression of myeloid preleukemia to transformed myeloid leukemia in children with down syndrome. Cancer Cell 2019;36:123-138.e10. [Crossref] [PubMed]
- Hyde RK, Kamikubo Y, Anderson S, et al. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood 2010;115:1433-43. [Crossref] [PubMed]
- Aivado M, Spentzos D, Germing U, et al. Serum proteome profiling detects myelodysplastic syndromes and identifies CXC chemokine ligands 4 and 7 as markers for advanced disease. Proc Natl Acad Sci U S A 2007;104:1307-12. [Crossref] [PubMed]
- Meldi K, Qin T, Buchi F, et al. Specific molecular signatures predict decitabine response in chronic myelomonocytic leukemia. J Clin Invest 2015;125:1857-72. [Crossref] [PubMed]
- Su C, Li H, Peng Z, et al. Identification of plasma RGS18 and PPBP mRNAs as potential biomarkers for gastric cancer using transcriptome arrays. Oncol Lett 2019;17:247-55. [PubMed]
- Dufies M, Giuliano S, Viotti J, et al. CXCL7 is a predictive marker of sunitinib efficacy in clear cell renal cell carcinomas. Br J Cancer 2017;117:947-53. [Crossref] [PubMed]


