
Highly expressed IFITM10 is associated with early diagnosis and T stage of gastric cancer
Introduction
Gastric cancer (GC) is the fifth most common malignant tumor that ranks third in mortality worldwide (1). Surgery is the main treatment for GC. However, most patients are already in advanced stage of the disease when they are diagnosed and cannot undergo radical surgery. Although the emerging programmed death 1 (PD-1) effectively improves patients survival in advanced GC, this effect is not completely satisfactory (2). Therefore, early GC screening has always been the most effective means of prevention and treatment. With the introduction of gastroscopy, early diagnosis of GC has made great progress. However, it remains of great significance to continue to develop effective diagnostic markers for early GC detection.
Interferon-induced transmembrane protein (IFITM) is widely present in mammals. Its functions include controlling cell proliferation, promoting homotypic cell adhesion, preventing viral infection, promoting bone matrix maturation and mineralization, and germ cell development (3). IFITM1 is highly expressed in colon cancer patients and can serve as an independent prognostic factor for colon cancer (4). IFITM2 is highly expressed in GC and is associated with its poor prognosis (5). IFITM3 is also highly expressed in esophageal cancer and can be used as a diagnostic marker for patients with stage IIA cancer (6). Highly expressed IFITM3 is closely associated with poor prognosis in acute leukemia (7). All genes in the IFITM family, except IFITM5, have a high expression after interferon stimulation (8). IFITM10 has the highest species conservation (9). The latest research shows that IFITM10 has weak antiviral properties (10). Fusion transcripts of IFITM10 and cathepsin D promote breast cancer proliferation (11). However, no direct correlation between IFITM10 expression level and tumors has been reported in the literature. Therefore, it is of great significance to explore the function of IFITM10, which will help to better understand the function of IFITM family proteins.
The present study analyzed transcriptome sequencing and clinical information on GC patients using the TCGA database and calculated the correlation between IFITM10 expression level and GC clinical phenotype and diagnosis. Surgical specimens and pathological information on GC patients were also collected during daily work to verify the above results. IFITM10 had a high expression in GC. This phenomenon was significantly associated with the T stage and diagnosis of GC. The potential IFITM10 function was also predicted using gene set enrichment analysis (GSEA) analysis.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2333).
Methods
Data mining
RNA-sequencing (RNA-Seq) expression (level 3 data) and clinical data for GC patients and normal tissues were collected form the TCGA database. RNA-Seq Expectation–Maximization (RSEM) expression values were used for statistical analysis. The expression level of IFITM10 in exosomes was obtained from exobase database (www.exorbase.org/).The construction of protein interaction network comes from string (string-db.org/).
Clinical samples
All clinical GC tissues and corresponding adjacent non-tumor samples were obtained from the Department of Gastrointestinal Nutrition and Hernia Surgery, the Second Hospital of Jilin University, Changchun, China. All samples were obtained during the operation and stored in RNA latter for further use (Invitrogen, Carlsbad, CA, USA) at −80 °C. None of the patients had chemotherapy and radiotherapy prior to surgery. All patients had a confirmed GC diagnosis using pathological analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Review Board of the Second Hospital of Jilin University [2018263] and informed consent was taken from all the patients.
RNA extraction and quantitative real-time PCR
Total RNA was extracted from GC tissues using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer instructions. For quantitative real-time PCR (qRT-PCR) detection, RNA was reverse transcribed to complementary DNA (cDNA) using a Reverse Transcription Kit (Takara, Dalian, China). The qPCR detection was performed using Roche 480 by SYBR II Premix Taq (Takara, Dalian, China). Ribosomal protein L13 (L13) was used as an internal reference (12). The specific primers were as follows: IFITM10 forward primer: GACGGGGCCTTCTGGATTC, IFITM10 reverse primer: TTCTTGTCTCGCACTTTGAGGGAG, L13 forward primer: CCAACGTGCAGCGGCCTGAA, and L13 reverse primer: CGTGGCCATCTTGAGTTCCT. Each sample was detected three times. The relative IFITM10 expression levels were compared to those of L13 and calculated using the 2-∆∆Ct method.
GSEA
In this study, GSEA was used to elucidate significant survival differences between high and low IFITM10 groups. The number of gene set permutations was 1,000 for each analysis. IFITM10 expression level was used as a phenotype label. Pathway enrichment was analyzed based on nominal P values and normalized enrichment score (NES).
Statistical analysis
All statistical analyses were conducted using R (v.3.6.1) (13) and SPSS version 22.0. Boxplots were used to visualize different expression levels via ggplot2 package in R. The relationship between clinical pathological features and IFITM10 was analyzed using the Wilcoxon signed-rank test and logistic regression. Kaplan-Meier curves were used to compare survival between high and low IFITM10 groups. ROC curves were generated using SPSS.
Results
Pathological GC patients information and IFITM10 expression level
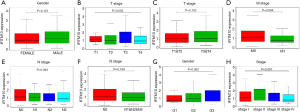
RNA-Seq data and corresponding tumor pathology information was collected for 375 GC and 32 normal tissues from the TCGA-STAD dataset. It included age, sex, survival, postoperative survival time, grade, T stage, N stage, M stage, and AJCC stage (Table 1). Patients with incomplete transcriptome sequencing and clinical information were excluded from the study. A total of 293 patients were included in the study. The two sets of data were analyzed using the Wilcoxon signed-rank test (Figure 1) or more groups by logistic regression (Table 2). IFITM10 mRNA was significantly highly expressed in tumor tissues in a data set containing 375 GC and 32 normal tissues (abnormally high expression values were eliminated, P<0.001, Figure 2). In addition, these tissues included 27 cases of GC and their matched normal tissues (abnormally high expression values were eliminated). IFITM10 had a higher expression in tumor tissues than in the paired tissues (P<0.001). Subsequently, the association between IFITM10 expression levels and clinical information was analyzed. IFITM10 had higher expression levels in tissues with a higher histological grade (P=0.062), in tissues with T1 stage (P=0.053), and in non-metastasized tumor tissues (P=0.084, Figure 2). Although these results have obvious trends, they are not statistically significant. Thus, surgical samples collected by the Second hospital of Jilin University were used for further result verification.
Table 1
| Characteristics | Number of cases | Percentages (%) |
|---|---|---|
| Gender | ||
| Male | 177 | 60.41 |
| Female | 116 | 39.59 |
| Histological grade | ||
| G1 | 5 | 1.71 |
| G2 | 102 | 34.81 |
| G3 | 186 | 63.48 |
| Topography (T) | ||
| T1 | 15 | 5.12 |
| T2 | 61 | 20.82 |
| T3 | 75 | 25.60 |
| T4 | 142 | 48.46 |
| Lymph node (N) | ||
| N0 | 92 | 31.40 |
| N1 | 76 | 25.94 |
| N2 | 66 | 22.53 |
| N3 | 59 | 20.14 |
| Metastasis (M) | ||
| M0 | 273 | 93.17 |
| M1 | 20 | 6.83 |
| AJCC stage | ||
| I | 41 | 13.99 |
| II | 94 | 32.08 |
| III | 126 | 43.00 |
| IV | 32 | 10.92 |
GC, gastric cancer; TCGA, the Cancer Genome Atlas.
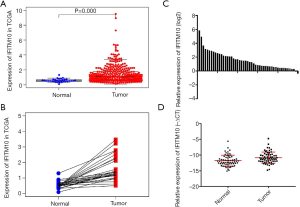
Table 2
| Characteristics | Total(N) | Odds ratio in IFITM10 expression | P value |
|---|---|---|---|
| T1 vs. T4 | 90 | 0.37 (0.11–1.15) | 0.097 |
| N0 vs. N3 | 151 | 0.62 (0.32–1.19) | 0.150 |
| M0 vs. M1 | 293 | 0.52 (0.19–1.31) | 0.180 |
| G1 vs. G3 | 191 | 1.63 (0.27–12.63) | 0.590 |
| I vs. IV | 73 | 0.52 (0.20–1.32) | 0.170 |
IFITM, Interferon-induced transmembrane.
The IFITM10 expression level was detected by qPCR in 62 pairs of GC tissues and their matched adjacent normal tissues (Table 3). The results showed that IFITM10 was highly expressed in GC tissues (P<0.001, Figure 1). Moreover, IFITM10 expression levels were higher in tissues with stages T1 and T2 (P=0.042), male patients (P=0.031), and tissues without neuro infiltration (P=0.008). These outcomes agree with previous results obtained from the TCGA.
Table 3
| Characteristics | Cases | Relative expression level of IFITM10 | P value |
|---|---|---|---|
| Sex | 0.031 | ||
| Male | 37 | 2.477 (1.368–5.618) | |
| Female | 25 | 1.392 (0.739–2.66) | |
| Age | 0.469 | ||
| ≤62 | 33 | 1.576 (1.225–3.227) | |
| >62 | 29 | 1.749 (1.376–4.648) | |
| Tumor size | 0.503 | ||
| ≤3.25 | 32 | 1.759 (1.344–4.38) | |
| >3.25 | 30 | 1.705 (0.865–3.171) | |
| T classification | 0.042 | ||
| T1 | 15 | 1.845 (1.502–4.313) | |
| T2 | 13 | 2.861 (1.488–5.414) | |
| T3 | 22 | 1.951 (0.887–4.361) | |
| T4 | 12 | 1.091 (0.504–1.638) | |
| T classification | 0.042 | ||
| T1&T2 | 28 | 2.322 (1.464–4.667) | |
| T3&T4 | 34 | 1.573 (0.573–2.955) | |
| N classification | 0.221 | ||
| N0 | 28 | 2.476 (1.388–5.554) | |
| N1 | 10 | 1.296 (0.708–1.65) | |
| N2 | 10 | 1.581 (1.315–3.326) | |
| N3 | 14 | 2.110 (0.887–4.026) | |
| N classification | 0.091 | ||
| N0 | 28 | 0.459 (0.232–1.332) | |
| N1&N2&N3 | 34 | 0.444 (0.107–0.876) | |
| M classification | 0.511 | ||
| M0 | 59 | 2.476 (1.388–5.554) | |
| M1 | 3 | 1.618 (0.86–2.955) | |
| AJCC stage | 0.210 | ||
| I | 21 | 2.861 (1.488–5.414) | |
| II | 13 | 1.674 (1.349–4.248) | |
| III | 22 | 1.539 (0.457–2.955) | |
| IV | 6 | 1.523 (1.285–2.263) | |
| Vascular invasion | 0.662 | ||
| Negative | 37 | 1.730 (1.271–4.38) | |
| Positive | 25 | 1.749 (0.739–3.498) | |
| Nerve infiltration | 0.008 | ||
| Negative | 49 | 2.294 (1.369–4.475) | |
| Positive | 13 | 0.739 (0.258–1.749) |
IFITM, Interferon-induced transmembrane; GC, gastric cancer.
These results indicated that IFITM10 was highly expressed in GC, especially in the early stages, suggesting that IFITM10 may be an effective diagnostic marker for early GC. The association between IFITM10 and patients survival and IFITM10 diagnostic potential were analyzed further.
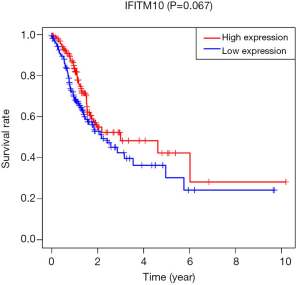
IFITM10 association with GC prognosis and diagnosis
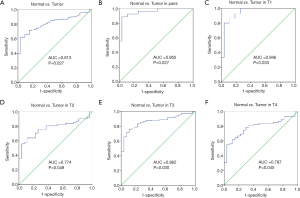
The survival package in R was used to map the survival curves for patients from the TCGA database. The results showed that patients with a high expression of IFITM10 had a longer survival (P=0.067, Figure 3). SPSS software was then used to construct the IFITM10 ROC curves based on the TCGA data. In all specimens, IFITM10 had an AUC value of 0.813, which suggested a moderate diagnostic potential. Among the 27 pairs of tissues, IFITM10 had an AUC value of 0.955 and an excellent diagnostic potential. The AUC in T1 stage tissues was 0.946, which suggested an excellent diagnostic potential (Figure 4). The diagnostic performance of IFITM10 is much higher than that of CEA and CA72-4 (14). We found that ifitm10 was highly expressed in exosomes from colon cancer, pancreatic cancer and liver cancer (Figure 5). Although there is no such study in GC. This indicated that IFITM10 could be used as a diagnostic marker for early GC. However, the impact of IFITM10 on prognosis requires more serum data to be confirmed.
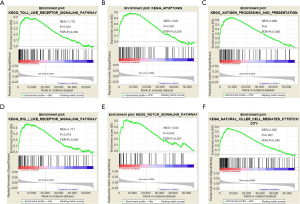
GSEA predicts IFITM10-related signaling pathways and protein-protein interactions
Because IFITM10 is a less reported gene in human cancer, its function was predicted using GSEA based on the NES and NOM-P-values to explain possible IFITM10 signaling pathways in GC. High expression of IFITM10 promoted antigen presentation and processing, natural killer cell-regulated cytotoxicity, RIGI-like receptor pathway, Notch pathway, toll-like receptor pathway, and apoptosis (Figure 6). Most of these pathways are related to tumor immunity. It was hypothesized that high expression of IFITM10 can activate the Notch signaling pathway, enhance the immune system, promote tumor cell death, and enhance the effect of tumor cells on immunotherapy. With the development of GC, IFITM10 expression decreased and resulted in loss of function, which may be the reason why some patients are tolerant to immunotherapy.
We constructed the protein interaction network of IFITM10 by string and listed top 10 interactant proteins (Figure 7). In these related proteins, the low expression of interferon regulatory factor 6 (IRF 6) is closely related to the poor prognosis of GC (15). Ubiquitin C-terminal hydrolase-L1 (UCHL1) can activate HIF-1α, which is a new tumor therapeutic target (16). SCNN1A is highly expressed in ovarian cancer and is closely related to the poor prognosis of ovarian cancer (17). IFITM10 can interact with these proteins and genes to participate in these tumor related pathways, which also shows that ifitm10 has certain research potential in cancer.
Discussion
The IFITM family of proteins is considered to be the first line of defense against human antiviral infections (18). There is a large number of studies on cancer-promoting effects of IFITM13 in tumors. In colon (4), breast (19), and lung cancers (20), IFITM1 is highly expressed and has a cancer-promoting effect. However, IFITM1 has a low expression in cervical squamous cell carcinoma and a tumor-suppressing effect, which is related to the high methylation level of IFITM1 in tumor tissues (21). In addition, IFITM1 is highly expressed in GC, but its high expression is associated with good prognosis for GC patients. This contradiction is believed to be related to the pathogenesis of tumors (22). IFITM3 is highly expressed in GC and has a cancer-promoting effect, but its expression decreases with increasing clinical stage (23). The present study experiments also found that IFITM10 had a similar unexplained expression trend.
IFITM protein family is overexpressed by interferon stimulation. However, gene level analysis found that IFITM10 is not on the conventional gene cluster for the IFITM family. Therefore, some researchers question whether IFITM10 is regulated by the interferon (9). However, the existing experiments only proved that IFN-α did not have the ability to activate IFITM10 (10). Some studies have shown that the level of interferon is lower in the serum of patients with GC compared to that in normal people. In addition, the level of interferon in GC patients is highest in stages I and IV, which may explain the abnormal expression of the partial IFITM family (24). Most of the current research focuses on IFITM13, in which gene positions of IFITM2 and 3 are extremely similar (9). In GC, they have a high expression and play a role in promoting cancer. However, IFITM2 gradually increases with GC progression (5). while IFITM3 has the opposite effect (5). This phenomenon also illustrates the complexity of the IFITM family of genes and their protein functions. In theory, the IFITM family generally promotes cell cycle arrest and apoptosis, which makes it seem that IFITM is a tumor suppressor gene. However, its actual expression and function are just the opposite. This may be due to epigenetics regulation or selective activation of gene function in a complex tumor environment. Although the present research showed that IFITM10 has an anti-cancer effect, it can be speculated that its activated downstream pathway is necessary for tumor survival. It is interesting that IFITM1 also showed high expression levels and tumor suppressive effects in GC. This completely opposite result from other tumors also suggested that GC may have unique biological characteristics, which are worthy of an in-depth consideration. Therefore, this result requires further in vivo and in vitro experiments to verify. Starting from IFITM10, it is of great significance to explore the function of the IFITM family.
Based on the GSEA analysis, IFITM10 may have an immune function that activates the body’s tumor cells. It may also be a sensitizer for immunotherapy, especially adoptive cellular immunotherapy (25). Immunotherapy is one of the current tumor biotherapy hotspots and some patients with advanced GC may get good results with this treatment. The development of immunotherapeutic sensitizers is crucial for the development of immunotherapy. Current research studies mainly utilize co-cultures with K562-mb15-4.1BBL cell line using ex vivo amplification to enhance the lethality of natural killer cells (26). In addition, the mechanism of tumor cell immune escape is mainly related to the lack of tumor cell surface antigen. IFITM10 function to enhance antigen presentation may partially reverse this phenomenon (27). An in-depth study of the IFITM10 function may help in the immunotherapy of GC. Notch signaling pathway plays a very complex role in tumors and can promote the process of epithelial mesenchymal transition (25). It also supports cancer-promoting effects in GC (28). Notch signaling pathway is currently an important target for cancer therapy. IFITM10 has a potential activation function for Notch, which means that IFITM10 may serve as a double-edged sword in tumors.
Some limitations were present in the study. First, due to the small number of paired specimens in the TCGA database, the experimental bias due to individual differences was not circumvented. The use of qPCR in patients specimens did not effectively measure the absolute expression of IFITM10, making it impossible to map a valid ROC. Second, the patients’ follow-up time was short and insufficient for survival analysis. Due to the lack of serum samples and relevant databases, we have not tested in this area. In addition, we did not have enough energy to perform the exact function verification of the new gene.
In conclusion, these experiments confirmed that IFITM10 was an effective marker for early diagnosis of GC. Its high expression represented the early T stage of GC and its function may be closely related to tumor immunity. IFITM10 was an intervention target with excellent potential.
Acknowledgments
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: This research was support by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2333
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2333
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2333
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2333). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Ethics Review Board of the Second Hospital of Jilin University (2018263) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Kamath SD, Kalyan A, Benson AB 3rd. Pembrolizumab for the treatment of gastric cancer. Expert Rev Anticancer Ther 2018;18:1177-87. [Crossref] [PubMed]
- Siegrist F, Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. J Interferon Cytokine Res 2011;31:183-97. [Crossref] [PubMed]
- He J, Li J, Feng W, et al. Prognostic significance of INF-induced transmembrane protein 1 in colorectal cancer. Int J Clin Exp Pathol 2015;8:16007-13. [PubMed]
- Xu L, Zhou R, Yuan L, et al. IGF1/IGF1R/STAT3 signaling-inducible IFITM2 promotes gastric cancer growth and metastasis. Cancer Lett 2017;393:76-85. [Crossref] [PubMed]
- Jia Y, Xiao Z, Jiang W, et al. Overexpression of IFITM3 predicts poor prognosis in stage IIA esophageal squamous cell carcinoma after Ivor Lewis esophagectomy. Thorac Cancer 2017;8:592-9. [Crossref] [PubMed]
- Liu Y, Lu R, Cui W, et al. High IFITM3 expression predicts adverse prognosis in acute myeloid leukemia. Cancer Gene Ther 2020;27:38-44. [Crossref] [PubMed]
- Zhang Z, Liu J, Li M, et al. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One 2012;7:e49265. [Crossref] [PubMed]
- Hickford D, Frankenberg S, Shaw G, et al. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genomics 2012;13:155. [Crossref] [PubMed]
- Okuzaki Y, Kidani S, Kaneoka H, et al. Characterization of chicken interferon-inducible transmembrane protein-10. Biosci Biotechnol Biochem 2017;81:914-21. [Crossref] [PubMed]
- Varley KE, Gertz J, Roberts BS, et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat 2014;146:287-97. [Crossref] [PubMed]
- Sharan RN, Vaiphei ST, Nongrum S, et al. Consensus reference gene(s) for gene expression studies in human cancers: end of the tunnel visible? Cell Oncol (Dordr) 2015;38:419-31. [Crossref] [PubMed]
- Team C. R: A Language and Environment for Statistical Computing. 2017.
- Rehena Z, Ghosh CK, Afroz F, et al. Comparison of Serum CA72-4 and CEA Levels in Patient with Endoscopically Suspected Gastric Carcinoma. Mymensingh Med J 2015;24:542-9. [PubMed]
- Li D, Cheng P, Wang J, et al. IRF6 Is Directly Regulated by ZEB1 and ELF3, and Predicts a Favorable Prognosis in Gastric Cancer. Front Oncol 2019;9:220. [Crossref] [PubMed]
- Nakashima R, Goto Y, Koyasu S, et al. UCHL1-HIF-1 axis-mediated antioxidant property of cancer cells as a therapeutic target for radiosensitization. Sci Rep 2017;7:6879. [Crossref] [PubMed]
- Wu L, Ling ZH, Wang H, et al. Upregulation of SCNN1A Promotes Cell Proliferation, Migration, and Predicts Poor Prognosis in Ovarian Cancer Through Regulating Epithelial-Mesenchymal Transformation. Cancer Biother Radiopharm 2019;34:642-9. [Crossref] [PubMed]
- Bailey CC, Zhong G, Huang IC, et al. IFITM-Family Proteins: The Cell's First Line of Antiviral Defense. Annu Rev Virol 2014;1:261-83. [Crossref] [PubMed]
- Lui AJ, Geanes ES, Ogony J, et al. IFITM1 suppression blocks proliferation and invasion of aromatase inhibitor-resistant breast cancer in vivo by JAK/STAT-mediated induction of p21. Cancer Lett 2017;399:29-43. [Crossref] [PubMed]
- Yan J, Jiang Y, Lu J, et al. Inhibiting of Proliferation, Migration, and Invasion in Lung Cancer Induced by Silencing Interferon-Induced Transmembrane Protein 1 (IFITM1). Biomed Res Int 2019;2019:9085435. [Crossref] [PubMed]
- Zheng W, Zhao Z, Yi X, et al. Down-regulation of IFITM1 and its growth inhibitory role in cervical squamous cell carcinoma. Cancer Cell Int 2017;17:88. [Crossref] [PubMed]
- Borg D, Hedner C, Gaber A, et al. Expression of IFITM1 as a prognostic biomarker in resected gastric and esophageal adenocarcinoma. Biomark Res 2016;4:10. [Crossref] [PubMed]
- Hu J, Wang S, Zhao Y, et al. Mechanism and biological significance of the overexpression of IFITM3 in gastric cancer. Oncol Rep 2014;32:2648-56. [Crossref] [PubMed]
- Zhao H, Dong N, Liu T, et al. Clinical Significance of Serum Type III Interferons in Patients with Gastric Cancer. J Interferon Cytokine Res 2019;39:155-63. [Crossref] [PubMed]
- Du Y, Wei Y. Therapeutic Potential of Natural Killer Cells in Gastric Cancer. Front Immunol 2019;9:3095. [Crossref] [PubMed]
- Mimura K, Kamiya T, Shiraishi K, et al. Therapeutic potential of highly cytotoxic natural killer cells for gastric cancer. Int J Cancer 2014;135:1390-8. [Crossref] [PubMed]
- Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res 2015;21:687-92. [Crossref] [PubMed]
- Demitrack ES, Samuelson LC. Notch as a Driver of Gastric Epithelial Cell Proliferation. Cell Mol Gastroenterol Hepatol 2017;3:323-30. [Crossref] [PubMed]



