
Circulating tumor DNA detection and its application status in gastric cancer: a narrative review
Introduction
Gastric cancer (GC) is the malignancy whose morbidity ranks the fourth place in the world. Up to 1 million new cases are diagnosed every year, and it is the second major cause of cancer-related death worldwide (1). China is one of the countries with the highest global morbidity of GC (2), with the 5-year survival rate of early GC of >90% and that of advanced GC of 30–40% (3). Therefore, the early diagnosis of GC is of particular importance to disease treatment and prognosis. For advanced GC, drug therapy remains the main treatment, and monitoring the drug responsiveness of tumors and timely discovering the occurrence of tumor resistance to avoid ineffective treatment is of crucial importance to extend the patient survival. Consequently, it is significant to search for a non-invasive technique with fewer traumas for the early diagnosis and dynamic monitoring of GC.
In 1984, Mandel and Metais (4) first reported the presence of DNA fragments in the blood; subsequently, they proposed the concept of cell-free DNA (cfDNA). Until 1994, Thierry et al. (5) and Diehl et al. (6) adopted the PCR technology to detect the mutant Ras proto-oncogene fragments in the plasma of pancreatic cancer and leukemia patients, respectively. Thus far, people have realized the importance of cfDNA. At present, the liquid biopsy mainly involves the detection of free circulating tumor cells (CTCs) in blood, circulating tumor DNA (ctDNA), circulating RNA and exosomes (carrying multiple cell source-related proteins, lipids, DNA and RNA). Among them, ctDNA, RNA and exosomes are secreted by the tumor itself, or released by the tumor cells at the time of death.
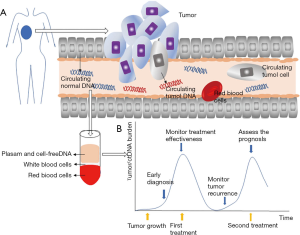
CtDNA is the small genomic fragment released by tumor cells into the circulating system (shown in Figure 1), which is characterized by the short half-life, carrying tumor-specific genetic/epigenetic variations, and great inter-individual difference. Besides, it is closely correlated with tumor development and evolution, dormancy and resistance, metastasis, and recurrence, and has gradually become the molecular research hotspot in the field of oncology. It is currently believed that ctDNA is derived from the primary tumor, metastasis, and CTCs. ctDNA carries the gene variation features, such as mutation, insertion, deletion, rearrangement, copy number variation (CNV) and methylation, rendering it an important biomarker (7). ctDNA has varied fragment size, which ranges from the small fragment of 70–200 bp to macromolecule of about 21,000 bp, and it is usually greater than the non-tumor cfDNA (8). Its number and property are affected by multiple factors, such as tumor burden, tumor status, DNA elimination, and degradation mechanism (9). In addition, the ctDNA level is also affected by the non-tumor-specific physiological or pathological mechanisms, such as inflammatory response or tissue injury (10). The half-life of ctDNA is about 2 h (11), therefore, it reflects the in vivo tumor status in a real-time manner. We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2856).
ctDNA detection
CtDNA detection includes quantitative detection and qualitative detection (Table 1). The former involves the blood/serum total copy number of ctDNA, while the latter involves the detection of tumor-specific genetic changes in blood/serum. ctDNA carrying tumor-related genomic and epigenomic changes represents the unique tumor information derived from patients. At present, numerous methods are available for detecting ctDNA, which are mainly classified as PCR- and next-generation sequence (NGS)-based methods, and each of them has its own merits and demerits, with no uniform standard so far. However, with the development of related technology, ctDNA will yield unusually brilliant results in the precision medicine of tumor.
Table 1
| Detection method | Specific detection content | Clinical significance of detection | References |
|---|---|---|---|
| Quantitative analysis of ctDNA | CtDNA expression level | (I) The high cpDNA level was related to the peritoneal recurrence and dismal prognosis for advanced GC patients | (12) |
| (II) The plasma cfDNA expression level is more sensitive to CEA, CA19-9 or CA50 in early GC screening, which may serve as the superior biomarker in GC screening | (13) | ||
| (III) The persistent existence or gradual increase in the blood HER2 amplification copy suggested the resistance-related tumor progression | (14) | ||
| Qualitative detection of ctDNA | Gene mutation | (I) Detecting the EGFR mutation in ctDNA is an effective method to identify patients who may benefit from the first-line gefitinib treatment | (11) |
| Gene rearrangement | (I) CtDNA analysis may be beneficial for patients when it is difficult to obtain the tissue sample | (15) | |
| Microsatellite instability (MSI) | (I) The ctDNA next-generation sequencing (ctDNA-NGS) might be used to detect the known MSI-H tumor patients | (16) | |
| DNA methylation | (I) RASSF1A and PCDH10 methylation in plasma ctDNA might serve as the potential non-invasive diagnostic index in GC blood | (17) | |
| (II) TIMP-3 methylation in body fluid was an independent risk factor for GC patients | (18) |
ctDNA, circulating tumor DNA; GC, gastric cancer.
Qualitative detection of ctDNA
Gene mutation
Under normal condition, oncogene exists in the form of proto-oncogene, which is at the non-carcinogenic status under the action of tumor suppressor gene. When the tumor suppressor gene is destroyed or mutated or the oncogene is mutated, the oncogene is activated to induce tumor genesis. However, the gene mutation rate is low. Additionally, the detectable mutated DNA level in peripheral blood is even lower, and ctDNA is easily lost during the DNA separation process due to its low content in peripheral blood. Moreover, considering the complexity of tumor gene variation and great individual differences, the detection of single gene mutation in clinical practice is greatly restricted. To sum up, these factors result in the low positive rate in peripheral blood ctDNA tumor mutant gene detection (8). Liao et al. (9) considered that these restrictions in ctDNA gene mutation detection lied in that they only focused on specific hotspot mutations with a limited number. In a study on hepatocellular carcinoma (HCC) conducted by Labgaa et al. (10), the authors adopted the superdeep sequencing of all exons to detect the HCC driver gene and medicable mutation (JAK1), which improved the previously reported plasma mutation detection rate in HCC patients. At the same time, the discovery of cis-mutation in plasma verified their tumor origin, which provided conclusive evidence that HCC-derived DNA fragments were released into the bloodstream. That study indicated the feasibility of superdeep sequencing for cfDNA, which might serve as a promising minimally invasive tool in HCC genetic detection. There are also reports on the clinical application of ctDNA gene mutation detection. For instance, a prospective clinical trial (11) used the ctDNA-based EGFR mutation detection as the criterion to select lung adenocarcinoma patients who received gefitinib as the first-line treatment. Besides, the NGS of 168 genomes is adopted for the genetic analysis of baseline blood samples. This study verifies that, detecting the EGFR mutation in ctDNA is an effective method to identify patients who may benefit from the first-line gefitinib treatment and that further analyzing the dynamic changes in EGFR mutation and the accompanying gene variations can predict the resistance to gefitinib.
Gene rearrangement
In a case report that used ctDNA to analyze the ALK fusion gene in colorectal cancer (CRC), Lai et al. (15) introduced a metastatic CRC patient who had not received treatment at the time of molecular detection. The patient was negative in the initial tumor tissue ALK immunohistochemical (IHC) staining, but the same STRN-ALK fusion gene was identified when a similar heterozygosis harvesting-based analytical method was utilized for parallel genome mapping analysis in ctDNA and tissues. Subsequently, the IHC staining was positive for ALK in the same samples, indicating that the initial result was false negative. Such result suggests that ctDNA analysis may be beneficial for patients when it is difficult to obtain the tissue sample.
Microsatellite instability (MSI)
Microsatellite is the region that contains multiple repeated 10–60 base pairs of 1–5 base pair sequences (19). Microsatellite occurs in the microsatellite locus, and it is extensively distributed in the entire human genome. In normal cells, the microsatellite repeat count is verified and maintained by the mismatch repair (MMR) system in the cellular DNA repair mechanism (20,21). Damage to the MMR system prevents cells from regulating its microsatellite length during the division period, which is referred to as MSI. After multiple cell division cycles, the damaged cells in the MMR system will form different lengths in their microsatellite sequences. MSI has been observed in CRC, endometrial cancer and gastric adenocarcinoma, and its clinical significance has been well described in CRC. For instance, CRC patients with high MSI (MSI-H) have been proved to have improved prognosis compared with tumor patients with microsatellite stability (MSS) (22,23). Maron et al. (16) discovered that, the ctDNA next-generation sequencing (ctDNA-NGS) might be used to detect the known MSI-H tumor patients. CtDNA-NGS can characterize the molecular heterogeneity of gastroesophageal adenocarcinoma and provide important prognosis and prediction information, and it serves as the supplement to tissue NGS. Moriyama et al. (24) detected the MSI and allele imbalance (AI) in plasma ctDNA from patients with pancreatic cancer and other biliopancreatic malignancies; meanwhile, they analyzed the five types of primary pancreatic cancers through microdissection and discovered that, the heterogeneity of the same pancreatic tumor was related to both MSI and AI. Thus, it may be an important marker of tumor heterogeneity, while plasma DNA analysis may become one of the diagnostic or therapeutic measures of such type of pancreatic cancer.
DNA methylation
Tumorigenesis is affected not only by genetic regulation, but also by epigenetic regulation (25). Epigenetic changes are one of the early events during the tumorigenesis process, including DNA methylation and histone modification. DNA methylation stands for the addition of the S-adenosine-L-methionine methyl into the cytosine or adenine ring of the CpG island, which plays an important role in silencing the tumor suppressor gene during the cancer development process (26,27). Pimson et al. (17) discovered that, the PCDH10 and RASSF1A gene methylation frequencies were high in GC tissues. The accumulate survival rates of RASSF1A methylation positive and (or) PCDH10 methylation positive patients were significantly lower than that in negative patients. Besides, high RASSF1A and PCDH10 methylation levels were discovered in advanced patients, which were significantly correlated with metastasis and histology. Therefore, the authors believed that, RASSF1A and PCDH10 methylation in plasma ctDNA might serve as the potential non-invasive diagnostic index in GC blood. Yu et al. (18) found that, the TIMP-3 gene methylation frequency in GC tissues was significantly higher than that in normal para-carcinoma tissues (P<0.001), and TIMP-3 methylation was tightly correlated with peritoneal metastasis and TNM stage (both P<0.001). In addition, Cox regression analysis revealed that, TIMP-3 methylation in body fluid was an independent risk factor for GC patients, and the disease-free survival (DFS) at 30 months after surgery significantly reduced.
Quantitative analysis of ctDNA
The landmark quantitative assessment of ctDNA in cancer was published by Leon et al. (28). They observed that metastatic patients showed higher cfDNA levels, which decreased as the clinical conditions of patients improved after radiotherapy. Fang et al. (12) discovered that, the median concentrations of circulating plasma DNA (cpDNA) in stage IV GC patients were apparently higher than those in stages I, II and III patients. Patients with high cpDNA levels were susceptible to peritoneal recurrence, with significantly lower 5-year overall survival (OS) than those with low cpDNA levels (P=0.039). Consequently, they believed that, the high cpDNA level was related to the peritoneal recurrence and dismal prognosis for advanced GC patients. Qian et al. (13) discovered significant differences in the plasma cfDNA expression level among 64 GC patients, benign gastric disease (BGD) patients and health controls (P<0.05). Among them, the plasma cfDNA expression in stage I GC patients was apparently higher than those in BGD patients and health controls (P<0.05), but no significant differences were observed in the traditional tumor markers CEA, CA19-9, CA72-4 and CA50 (P>0.05). It is indicated that, the plasma cfDNA expression level is more sensitive to CEA, CA19-9 or CA50 in early GC screening, which may serve as the superior biomarker in GC screening. Wang et al. (14) proposed in a study examining the resistance mechanism of Trastuzumab that, the ctDNA-based HER2 amplification transcript might be used to determine the resistance of HER2+ GC patients to Trastuzumab treatment. They conducted ctDNA atlas analysis on 97 series plasma samples from 24 HER2+ GC patients receiving Trastuzumab combined with chemotherapy, and found that the resistant cloning might be produced through changing the HER2 amplification cloning to overcome the Trastuzumab treatment, besides, the persistent existence or gradual increase in the blood HER2 amplification copy suggested the resistance-related tumor progression.
Clinical significance of ctDNA detection in advanced GC (Table 2)
Table 2
| Cancer screening | Early diagnosis |
|---|---|
| Early GC | Identify specific genomic changes to guide treatment selection |
| Detection of MRD | |
| Monitor treatment effectiveness | |
| Assess the prognosis | |
| Advanced GC | Monitoring GC metastasis and recurrence |
| Guide the choice of treatment | |
| Monitor treatment effectiveness | |
| Assess the prognosis | |
| Refractory GC | Discover new genome changes |
| Study drug resistance mechanism | |
| Guide the choice of treatment |
ctDNA, circulating tumor DNA; GC, gastric cancer; MRD, minimal residual disease.
Early diagnosis
Specific clinical manifestations are lacking in early GC, which can hardly be discovered and is usually diagnosed at an advanced stage, leading to its low 5-year survival of only 30–40%. However, the survival rate of early GC significantly increases (3). Tumor has secreted ctDNA into the blood before the disease sign is detected by imaging examination and (or) detection, so ctDNA may serve as one of the most promising biomarkers in early cancer detection (29).
Multiple studies have discovered the different plasma ctDNA levels between tumor patients and healthy subjects. Some research discovers that (30,31) the plasma cfDNA levels in GC patients are apparently higher than those in healthy subjects, and those in advanced GC patients are higher than those in early GC cases. At the same time, research indicates that, the ctDNA detection rate in multiple advanced tumor patients is as high as 85%, but that is relatively low in early tumor patients (32). Cohen et al. (33) developed an early cancer detection test named “CancerSEEK”, which mainly targeted cancers that were hardly screened. They used a set of 61 amplicons to analyze the blood samples, so as to detect the common mutations in several solid tumor subtypes (including breast, colorectal, esophageal, liver, lung, ovarian, pancreatic and GCs), and they were blinded to the specific mutations of these tumors. Another set of 8 common protein cancer biomarkers [cancer antigen 125 (CA-125), carcino-embryonic antigen (CEA), cancer antigen 19-9 (CA 19-9), hepatocyte growth factor (HGF), myeloperoxidase (MPO), osteopontin (OPN), prolactin (PRL) and tissue inhibitor of metalloproteinase-1 (TIMP-1)] further improved the detection rate. Their test achieved a 99% specificity, and only 7 out of the 812 health controls had false positive results, while the sensitivity ranged from 33% of breast cancer to 98% of ovarian cancer. On the whole, the authors determined the cancer-specific detection profile that might be used in the early detection (stages I–III) of over 82% of cancers. One major finding of that study was that, in some indiscoverable cancers like ovarian cancer, liver cancer, pancreatic cancer, esophageal cancer and GC, the early discovery rate was 69% or higher. Nonetheless, no screening item is recommended for these cancers in related guidelines.
Prognosis assessment
After treatment for gastrointestinal tract malignancy, ctDNA may become the potential biomarker of minimal residual disease (MRD). The detection of ctDNA may indicate a higher risk of recurrence in patients, even though there is no clinical evidence for other diseases (34). Normando (35) observed that, after the first chemotherapy cycle (3 months), patients with low ctDNA levels had significantly prolonged DFS compared with those with high ctDNA levels (COX regression P=0.0228). At 3 months after chemotherapy in advanced GC patients, the ctDNA level was positively correlated with DFS. Fang et al. (12) verified that, the high ctDNA level was related to peritoneal recurrence and poor prognosis in advanced GC patients, and advanced GC patients with ctDNA mutation had a dismal prognosis. Dardaei et al. (36) discovered that the OS of GC patients was significantly correlated with SOX17 methylation (P=0.049). Meanwhile, the methylation status was also significantly correlated with the differentiation level (P=0.031). In the free DNA of resectable GC patients, the methylation of Sox17 gene promoter is a common event, which may provide important information to judge the patient prognosis. In a meta-analysis including 16 studies and 1193 GC patients, the presence of ctDNA was significantly related to the short DFS and OS in GC patients, which achieved relatively high specificity and moderate sensitivity (37). Han et al. (38) also discovered that, the MINT2 gene methylation status in tumor tissues was apparently higher than that in normal para-carcinoma tissues. The methylated MINT2 level in tumor tissues was extremely close to those in paired preoperative peritoneal lavage fluid and blood samples. This might be because the abnormal methylation of MINT2 in tumor tissue was tightly related to the peritoneal dissemination, tumor progression and dismal prognosis.
Recurrence monitoring
CtDNA has a short half-life, which is rapidly eliminated from the plasma. Therefore, ctDNA detection can more accurately monitor the tumor dynamic and load changes during the course of treatment, and it can usually discover recurrence earlier than traditional imaging examination.
Pu et al. (39) discovered in a longitudinal study that, the ctDNA levels elevated before surgery and at 21 days after surgery, but declined at 3 months after surgery, and elevated again in the case of disease progression. Lan et al. (40) discovered in a large study on 428 GC patients that, the persistently high ctDNA level after surgical resection was an indicator of GC recurrence. Kim et al. (41) verified that, the median duration (namely, median lead time) from positive ctDNA detection to recurrence was 4.05 months. In addition, the postoperative positive ctDNA was significantly correlated with tumor recurrence within 12 months after surgery (P=0.029), while preoperative positive ctDNA was not related to tumor recurrence. Such result reveals that postoperative ctDNA monitoring is of certain clinical application value in the prediction of postoperative GC recurrence.
Therapeutic effect monitoring
In a study examining the resistance mechanism of Trastuzumab (14), the ctDNA-based HER2 amplification transcript is proposed to be used to monitor the resistance of HER2+ GC patients to Trastuzumab treatment. It is discovered that, the persistent existence or gradual increase in the blood HER2 amplification copy suggests the resistance-related tumor progression.
Clinical research proves the therapeutic effect of programmed cell death 1 (PD-1) targeted therapy on some metastatic gastric cancer (MGC) patients. To determine the determinant of response, Kim et al. (42) carried out molecular characterizations on tissue and ctDNA from 61 MGC patients receiving pembrolizumab treatment in a prospective phase II clinical trial. After 6 weeks of treatment, the changes in ctDNA levels predicted the response and DFS, and ctDNA decrease was related to the improved outcomes. They believed that, with regard to the biomarker to monitor the pembrolizumab therapeutic effect, ctDNA mutation load might serve as a feasible choice, or at least a supplementary testing tool. Chen et al. (43) aimed to quantify chromosome instability by the ctDNA copy number instability (CNI) during the drug therapy course for advanced gastric cancer (AGC), and subsequently to assess the correlation of chromosome instability with the therapeutic response. They discovered that, the ctDNA-based chromosome instability might be used to predict and monitor the therapeutic response for GC.
Prospects
With the development of tumor precision therapy, it is urgently needed to develop a tumor biomarker with higher sensitivity and specificity to guide clinical practice. The tumor genome information carried by ctDNA is consistent with that by tumor tissues, thus, ctDNA detection can overcome the unbreakable tumor heterogeneity problem encountered in conventional tumor tissue biopsy. Moreover, the detection method is simple, real-time, and non-invasive, which has thereby attracted increasing attention. This review indicates that, the ctDNA detection research can be conducted from several aspects at present, including quantitative detection, gene mutation, gene rearrangement, microsatellite instability and gene methylation, which is of great significance to the early diagnosis, prognosis evaluation, disease monitoring and individualized treatment for GC.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2856
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2856). HY reports grants from Key Program of the Changzhou Commission of Health, during the conduct of the study. XL reports grants from Young Talents of the Changzhou Commission of Health, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bertuccio P, Chatenoud L, Levi F, et al. Recent patterns in gastric cancer: a global overview. Int J Cancer 2009;125:666-73. [Crossref] [PubMed]
- Wang J, Yu JC, Kang WM, et al. Treatment strategy for early gastric cancer. Surg Oncol 2012;21:119-23. [Crossref] [PubMed]
- Strong VE, D'Amico TA, Kleinberg L, et al. Impact of the 7th Edition AJCC staging classification on the NCCN clinical practice guidelines in oncology for gastric and esophageal cancers. J Natl Compr Canc Netw 2013;11:60-6.
- Mandel P, Metais P. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil 1948;142:241-3. [PubMed]
- Thierry AR, Mouliere F, Gongora C, et al. Origin and quantification of circulating DNA in mice with human colorectal cancer xenografts. Nucleic Acids Res 2010;38:6159-75. [Crossref] [PubMed]
- Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. [Crossref] [PubMed]
- Esposito A, Criscitiello C, Locatelli M, et al. Liquid biopsies for solid tumors: Understanding tumor heterogeneity and real time monitoring of early resistance to targeted therapies. Pharmacol Ther 2016;157:120-4. [Crossref] [PubMed]
- Qi Q, Pan YF, Shen JJ, et al. Circulating DNA for detection of gastric cancer. Eur Rev Med Pharmacol Sci 2016;20:2558-64. [PubMed]
- Liao W, Yang H, Xu H, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016;7:40481-90. [Crossref] [PubMed]
- Labgaa I, Villacorta-Martin C, D'Avola D, et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 2018;37:3740-52. [Crossref] [PubMed]
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018;6:681-90. [Crossref] [PubMed]
- Fang WL, Lan YT, Huang KH, et al. Clinical significance of circulating plasma DNA in gastric cancer. Int J Cancer 2016;138:2974-83. [Crossref] [PubMed]
- Qian C, Ju S, Qi J, et al. Alu-based cell-free DNA: a novel biomarker for screening of gastric cancer. Oncotarget 2016;8:54037-45. [Crossref] [PubMed]
- Wang DS, Liu ZX, Lu YX, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut 2019;68:1152-61. [Crossref] [PubMed]
- Lai AZ, Schrock AB, Erlich RL, et al. Detection of an ALK Fusion in Colorectal Carcinoma by Hybrid Capture-Based Assay of Circulating Tumor DNA. Oncologist 2017;22:774-9. [Crossref] [PubMed]
- Maron SB, Chase LM, Lomnicki S, et al. Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma. Clin Cancer Res 2019;25:7098-112. [Crossref] [PubMed]
- Pimson C, Ekalaksananan T, Pientong C, et al. Aberrant methylation of PCDH10 and RASSF1A genes in blood samples for non-invasive diagnosis and prognostic assessment of gastric cancer. PeerJ 2016;4:e2112. [Crossref] [PubMed]
- Yu JL, Lv P, Han J, et al. Methylated TIMP-3 DNA in body fluids is an independent prognostic factor for gastric cancer. Arch Pathol Lab Med 2014;138:1466-73. [Crossref] [PubMed]
- Schlötterer C. Genome evolution: are microsatellites really simple sequences? Curr Biol 1998;8:R132-4. [Crossref] [PubMed]
- Shia J. Evolving approach and clinical significance of detecting DNA mismatch repair deficiency in colorectal carcinoma. Semin Diagn Pathol 2015;32:352-61. [Crossref] [PubMed]
- Strand M, Prolla TA, Liskay RM, et al. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 1993;365:274-6. [Crossref] [PubMed]
- Buckowitz A, Knaebel HP, Benner A, et al. Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer 2005;92:1746-53. [Crossref] [PubMed]
- Benatti P, Gafa R, Barana D, et al. Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res 2005;11:8332-40. [Crossref] [PubMed]
- Moriyama H, Matsubara N, Kanbara T, et al. Allelic imbalance and microsatellite instability in plasma DNA released from polyclonal pancreatic adenocarcinoma. Int J Oncol 2002;21:949-56. [Crossref] [PubMed]
- Gold B, Cankovic M, Furtado LV, et al. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J Mol Diagn 2015;17:209-24. [Crossref] [PubMed]
- Levenson VV. DNA methylation as a universal biomarker. Expert Rev Mol Diagn 2010;10:481-8. [Crossref] [PubMed]
- Warton K, Samimi G. Methylation of cell-free circulating DNA in the diagnosis of cancer. Front Mol Biosci 2015;2:13. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Campos-Carrillo A, Weitzel JN, Sahoo P, et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol Ther 2020;207:107458. [Crossref] [PubMed]
- Kim K, Shin DG, Park MK, et al. Circulating cell-free DNA as a promising biomarker in patients with gastric cancer: diagnostic validity and significant reduction of cfDNA after surgical resection. Ann Surg Treat Res 2014;86:136-42. [Crossref] [PubMed]
- Park JL, Kim HJ, Choi BY, et al. Quantitative analysis of cell-free DNA in the plasma of gastric cancer patients. Oncol Lett 2012;3:921-6. [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Cohen JD, Li L, Wang Y, et al. Detection and localization of surgically resec cancers with a multi-analyte blood test. Science 2018;359:926-30. [Crossref] [PubMed]
- Sawada K, Kotani D, Bando H. The Clinical Landscape of Circulating Tumor DNA in Gastrointestinal Malignancies. Front Oncol 2018;8:263. [Crossref] [PubMed]
- Normando SRC, Delgado PO, Rodrigues A, et al. Circulating free plasma tumor DNA in patients with advanced gastric cancer receiving systemic chemotherapy. BMC Clin Pathol 2018;18:12. [Crossref] [PubMed]
- Dardaei L, Shahsavani R, Ghavamzadeh A, et al. The detection of disseminated tumor cells in bone marrow and peripheral blood of gastric cancer patients by multimarker (CEA, CK20, TFF1 and MUC2) quantitative real-time PCR. Clin Biochem 2011;44:325-30. [Crossref] [PubMed]
- Gao Y, Zhang K, Xi H, et al. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: a meta-analysis. Oncotarget 2017;8:6330-40. [Crossref] [PubMed]
- Han J, Lv P, Yu JL, et al. Circulating methylated MINT2 promoter DNA is a potential poor prognostic factor in gastric cancer. Dig Dis Sci 2014;59:1160-8. [Crossref] [PubMed]
- Pu WY, Zhang R, Xiao L, et al. Prediction of cancer progression in a group of 73 gastric cancer patients by circulating cell-free DNA. BMC Cancer 2016;16:943. [Crossref] [PubMed]
- Lan YT, Chen MH, Fang WL, et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget 2017;8:3009-17. [Crossref] [PubMed]
- Kim YW, Kim YH, Song Y, et al. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp Mol Med 2019;51:1-10. [Crossref] [PubMed]
- Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449-58. [Crossref] [PubMed]
- Chen Z, Zhang C, Zhang M, et al. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis 2019;10:697. [Crossref] [PubMed]


