
Microarray analysis of miRNA based on the regional lymph node metastasis status of esophageal squamous cell carcinoma
Introduction
Esophageal cancer (EC) is one of the most common malignant tumors in the world, ranking eighth in cancer-related morbidity and sixth in cancer-related mortality worldwide (1). Esophageal squamous cell carcinoma (ESCC) is the dominant histological subtype in EC, accounting for almost 90% of EC patients, especially in China (2). For decades, although researchers have endeavored to find breakthroughs in the treatment of ESCC, the outcome has been disappointing and the survival rate remains dismal (3,4). One of the most important reasons for the failure of ESCC treatment is regional lymph node metastasis (LNM); the regional LNM rate in ESCC is reported to reach 40–70% (5,6). In ESCC, the lymphatic drainage regions include the lower neck, supraclavicular, mediastinum, and even the upper abdomen. It is very difficult to control such a wide range of lymphatic drainage regions for surgery and radiotherapy, which are the two main treatments of ESCC, because the control of such a wide range of lymphatic drainage regions will greatly increase the difficulty and complication of treatment, even might lead to the death of patients (7-9). Therefore, the control of a selective range of lymphatic drainage regions will be conducive to reduce the difficulty and complication of treatment, and will be worthy of be studied.
In fact, although in clinical practice there is a relatively high and wide incidence of LNM in ESCC, there is a great difference among patients (5,6). In current clinical practice, in order to distinguish this difference, lymph node dissection or irradiation is usually carried out selectively according to the location of ESCC (10). However, due to the characteristics of lymph node jumping in the metastasis of ESCC (11), these disposals often lead to the omission and increase the risk of LNM after treatment. With the progression of modern molecular biology, researchers are gradually realizing that intrinsic molecular biological characteristics may lead to the great difference in LNM among different patients (12). Therefore, the search for an appropriate molecular biomarker to predict regional LNM to guide the selective lymphatic dissection or irradiation of ESCC will undoubtedly reduce the difficulty and complication of treatment, decrease the incidence of treatment failure for LNM, increase survival rate of ESCC, and bring great progress in the treatment of ESCC.
MicroRNAs (miRNAs) are a class of non-coding small RNAs, which can modulate the target gene expression at the post-transcriptional level. MiRNAs participate in a variety of physiological and pathological processes, including cell proliferation, cell death, stress response, apoptosis, fat metabolism, cell differentiation, and so on (13). Accumulating evidence has shown that miRNAs are closely associated with malignant tumors, and many miRNAs act as oncogenes or tumor suppressors in several malignant tumors (14). In recent years, some studies have found that some miRNAs participate in the regulation of cancer invasion and metastasis, and can be used for the prediction of cancer invasion and metastasis (15,16). In ESCC, previous studies have found that miRNA-145, miRNA-200b, and miRNA-208 are involved in regulating the invasion and metastasis of ESCC, and miRNA-17a, miRNA-19b, miRNA-302b, miRNA-100, miRNA-199a, and miRNA-92a were associated with LNM in ESCC (17,18). However, these conclusions are mostly based on experiments in vitro or with a small sample of clinical cases. Due to the lack of characteristic studies based on the regional LNM status of ESCC, there are no miRNAs that can be used for the prediction of regional LNM in ESCC at present.
Therefore, in this study, mature miRNA microarray analysis based on regional LNM status was used to screen for miRNAs with significant differential expression in ESCC. Simultaneously, the verification of a large sample of ESCC tissues with or without regional LNM was performed to obtain tumor-specific miRNAs for the prediction of regional LNM in ESCC to guide the clinical treatment of ESCC. We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-1969).
Methods
Samples
Primary tumor tissues were taken from EC patients and immediately frozen in liquid nitrogen after surgical resection and prepared for the experiments. All the samples selected were in accord with the following criteria: patients underwent radical EC resection and regional lymphatic dissection; samples were pathologically confirmed as ESCC postoperatively and regional LNM status was pathologically evaluated postoperatively; none of preoperative chemotherapy or radiotherapy was taken; all the patients did not accompanied with any other serious diseases and the expected survive was more than 6 months. Eligible patients were registered from the Shandong Cancer Hospital and the Second Hospital of Shandong University from September 2017 to March 2018. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All the patients signed the informed consent, and the study was approved by the Ethics Committees of the two hospitals [registration number: KYLL-2017(GJ)P-0036].
RNA isolation and microarray analysis
According to the manufacturer’s protocol, total RNA was respectively isolated from four pairs of ESCC tissues with and without LNM using a TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and then was purified using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). To ensure that the total RNA isolated met the experimental requirements, a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific) was used to quantify the purity and concentration of total RNA, and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) was used to evaluate the integrity of total RNA.
After verifying that the quality of total RNA extraction from four pairs of ESCC tissues reached the experimental requirements, miRNA expression profiles of ESCC tissue with and without LNM were analyzed with the human miRNA array version 21.0 (Agilent). During the miRNA array analysis, a miRNA Complete Labeling and Hyb Kit (Agilent) was firstly used to label miRNAs according to the manufacturer’s protocol. Then the Cy3-labeled samples were hybridized according to the guidelines for the Agilent miRNA array. Through the whole miRNA array analysis, the labeling spike-in RNA and the Hyb spike-in RNA from Agilent were used for the quality control of the experimental process. After the hybridization, the slides were washed and dried according to the manufacturer’s manuals, and then were scanned using the Agilent microarray scanner.
Following the scan, the images were imported into the Agilent Feature Extraction software (v10.7) (Agilent Instruments) for further grid alignment and data extraction. After that, Agilent GeneSpring software was used for the normalization of the raw signal data extracted. And then, the differentially expressed miRNAs between two groups were identified using a standard fold change (abs) >2 and P<0.05. The entire miRNAs datasets mentioned here are available from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/)
miRNA target gene prediction analysis
After microarray analysis, miRNA target gene prediction analysis was performed for the differentially expressed miRNAs. MiRNA target gene prediction analysis using miRWalk2.0 (http://zmf.umm.uni-heidelberg.de/mirwalk2/) generated the prediction results of miRNA target genes from 12 existing miRNA-target prediction programs, including DIANA-microTv4.0, DIANA-microT-CDS, miRanda-rel2010, mirBridge, miRDB4.0, miRmap, miRNAMap, PicTar2, PITA, RNA22v2, RNAhybrid2.1, and Targetscan6.2. A gene would be regarded as a target of a differentially expressed miRNA if it was predicted in at least seven of the 12 miRNA-target prediction programs.
Gene ontology enrichment and pathway analysis
After miRNA target gene prediction analysis, gene ontology (GO) enrichment and pathway analyses of the target mRNAs for the differentially expressed miRNAs were performed. The GO project provides the annotations for the genes and gene products of any organism (http://www.geneontology.org), which covers the roles of mRNAs in the three domains including biological process, cellular component, and molecular function. The Fisher’s exact method was used to test whether the overlap between the differentially expressed list and the GO annotated list was greater than that expected by chance. While pathway analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis was used to explore the function of target genes. The Fisher’s exact test was used to identify a significant pathway, and the P value was used to defined the threshold of significance; the lower P value represented the more significant pathway.
MiRNA screen and verification
Based on the results of the microarray analysis, the differentially expressed miRNAs were screened for verification according to the criteria as follow: (I) fold change >2.0 and P<0.05, and (II) the raw signal intensity of each miRNA >1 and with a “detected” Feature Extraction Flag. After the miRNA screen, 50 ESCC samples, including 25 without LNM and 25 with LNM were selected for miRNA verification using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The significant miRNAs were regarded as candidate miRNAs capable of predicting regional LNM in ESCC.
Correlation analysis between candidate miRNAs and clinicopathologic features
To verify the specificity of the candidate miRNAs capable of predicting regional LNM in ESCC, the relations between verified miRNAs and clinicopathologic features were also analyzed. The clinicopathologic features analyzed included sex, age, tumor size, tumor location, T stage, TNM stage, tumor clinical subtype and tumor differentiation degree. Finally, tumor-specific miRNAs capable of predicting regional LNM in ESCC were obtained. We defined the postoperative stage according to the 2017 version TNM staging classification system of the American Joint Committee on Cancer (AJCC).
Potential mechanism analysis of the specific miRNAs
According to the results of the miRNA target gene prediction analysis, potential target genes of the tumor-specific miRNAs and their enriched pathways were also analyzed, and the potential mechanisms of the tumor-specific miRNAs regulating regional LNM in ESCC were predicted.
Statistical analysis
Statistical analysis was carried out using SPSS 20.0 software (IBM SPSS Statistics for Windows, Armonk, NY, USA) except the analysis methods indicated above. The expression levels of miRNAs were displayed as the mean ± standard deviation (SD). The expression differences of miRNAs between the two groups were compared with the independent-samples t-test. The differences in clinicopathologic features between the two groups were compared using the chi-square test. The correlations between specific miRNAs and tumor size were tested with Pearson correlation analysis. The value of P<0.05 was considered statistically significant.
Results
Sample selection
According to the inclusion criteria of samples, except LNM status, the clinicopathologic features of the four pairs of ESCC tissues selected for microarray analysis were matched, and those of the 50 tumor samples including 25 without LNM and 25 with LNM used to verify the effectiveness of the screened miRNAs were similar between the two groups (all P values >0.05). The clinicopathologic features of the samples selected in this study are presented in Tables 1,2.
Table 1
| Sample | Age (years) | Gender | Primary location | Pathology grade | Clinical subtype | Tumor size (cm2) | Stages† |
|---|---|---|---|---|---|---|---|
| NLNM‡1 | 59 | Male | Lower thoracic esophagus | Moderate differentiation | Ulcerative type | 5.8×2.5 | pT3N0M0 |
| NLNM2 | 62 | Male | Midthoracic esophagus | Moderate differentiation | Ulcerative type | 6.0×2.3 | pT2N0M0 |
| NLNM3 | 68 | Male | Lower thoracic esophagus | Poor differentiation | Ulcerative type | 5.0×1.6 | pT3N0M0 |
| NLNM4 | 67 | Female | Lower thoracic esophagus | Moderate differentiation | Ulcerative type | 6.5×3.0 | pT3N0M0 |
| LNM§1 | 58 | Male | Lower thoracic esophagus | Moderate differentiation | Ulcerative type | 5.0×3.0 | pT3N3M0 |
| LNM2 | 73 | Male | Midthoracic esophagus | Moderate differentiation | Ulcerative type | 4.0×3.5 | pT2N1M0 |
| LNM3 | 75 | Male | Lower thoracic esophagus | Poor differentiation | Ulcerative type | 3.0×2.5 | pT3N2M0 |
| LNM4 | 56 | Female | Lower thoracic esophagus | Moderate differentiation | Ulcerative type | 5.0×4.0 | pT3N1M0 |
†, postoperative pathological stage; ‡, NLNM: represent sample without lymph node metastasis; §, LNM: represent sample with lymph node metastasis.
Table 2
| Main features | Group 1† | Group 2‡ | P value |
|---|---|---|---|
| Gender (n) | 1.000 | ||
| Male | 20 | 20 | |
| Female | 5 | 5 | |
| Median age (year) | 62 (46 to 76) | 63 (45 to 82) | |
| Age (n) | 0.564 | ||
| ≥60 years | 14 | 16 | |
| <60 years | 11 | 9 | |
| Primary location (n) | 0.258 | ||
| Midthoracic esophagus | 14 | 10 | |
| Lower thoracic esophagus | 11 | 15 | |
| Pathology grade (n) | 0.248 | ||
| Poor | 8 | 12 | |
| Moderate and well | 17 | 13 | |
| Clinical subtype (n) | 0.395 | ||
| Ulcerative type | 10 | 13 | |
| Medullary and other type | 15 | 12 | |
| Tumor size (mean ± SD) (cm2) | 8.14±5.55 | 9.21±6.59 | 0.539 |
| T stage (n) | 0.480 | ||
| T1–2 | 6 | 4 | |
| T3 | 19 | 21 | |
| N stage (n) | – | ||
| N0 | 25 | 0 | |
| N1 | 0 | 18 | |
| N2 | 0 | 6 | |
| N3 | 0 | 1 | |
| TNM stage§ (n) | – | ||
| IB | 1 | 0 | |
| IIA | 14 | 0 | |
| IIB | 10 | 1 | |
| IIIA | 0 | 3 | |
| IIIB | 0 | 20 | |
| IVA | 0 | 1 | |
†, Group 1: expression levels of miRNAs in cancer tissues without lymph node metastasis; ‡, Group 2: expression levels of miRNAs in cancer tissues with lymph node metastasis; §, postoperative pathological stage.
Overview of the microarray analysis
To guarantee the reliability of the microarray analysis results, strict quality control was performed in the sample selection, storage to RNA extraction, and microarray analysis. The final results of the microarray analysis reached our expectations.
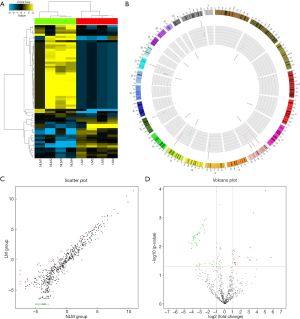
According to the results of the comparative microarray analysis, 62 miRNAs showed obvious differential expression (fold change ≥2.0, P<0.05) in ESCC samples with LNM compared to those without LNM. Among them, 19 miRNAs were upregulated and 43 miRNAs were downregulated in ESCC samples with LNM compared to those without LNM. The results are expressed as a cluster plot, scatter plot, volcano plot, and Circos plot in Figure 1.
MiRNA target gene prediction analysis
Further miRNA target gene prediction analysis identified 50 miRNAs, including 19 upregulated miRNAs and 31 downregulated miRNAs, among the 62 differentially expressed miRNAs with related target genes (target mRNA) according to the 12 existing miRNA-target prediction programs based on miRWalk2.0. The number of predicted target genes reached 8,637 and the potentially related pairs of miRNA-mRNA reached 37,231 according to the miRNA target gene prediction criteria.
GO and pathway analyses
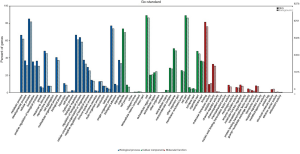
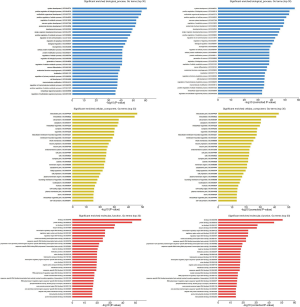
To understand the functions of the underlying target mRNAs, GO analysis and pathway analysis were performed. The results of GO analysis showed that the majority of underlying target mRNAs was involved in biological process, one fraction of them was defined as cellular component, and a few of them participated in molecular function (these are shown in Figure 2). Among the roles of mRNAs, cellular processes, single-organism processes, metabolic processes, biological regulation, and regulation of biological processes were the top five biological processes involved in the greatest number of potential target mRNA. Cell, cell parts, and organelles were the top three cellular components involved in the greatest number of potential target mRNA. While binding and catalytic activity were the top two molecular functions involving the greatest number of potential target mRNA. The top 30 significantly enriched GO terms are also shown in Figure 3.
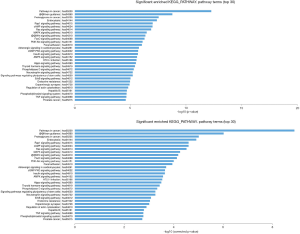
In the KEGG pathway analysis, the top 30 significantly enriched KEGG pathways are shown, and among them, pathways in cancer, proteoglycans in cancer, Rap1 signaling pathway, Ras signaling pathway, MAPK signaling pathway, Wnt signaling pathway, PI3K-Akt signaling pathway, ErbB signaling pathway, and TNF signaling pathway were the significant signaling pathways associated with the pathogenesis of cancer and involved in potential target mRNAs. The results are shown in Figure 4.
miRNA screen and qRT-PCR verification
According to the screening criteria for miRNAs and the attempt to verify more miRNAs, 19 miRNAs, including 11 upregulated and eight downregulated miRNAs, were selected among the differentially expressed miRNAs for further qRT-PCR verification. In the verification, 50 ESCC samples, including 25 without LNM and 25 with LNM were selected to verify the effectiveness of the screened miRNAs. The results of qRT-PCR verification showed that the expression tendency of 14 miRNAs was consistent with that of the microarray data, and the consistency was 73.68%. Among the 19 miRNAs verified, five miRNAs were significantly upregulated, no miRNAs were significantly downregulated in ESCC tissues with LNM compared with those without LNM. Among the five significantly upregulated miRNAs, the expression of miR-4440 and miR-6127 were inconsistent between the qRT-PCR verification data and microarray data, and they were therefore excluded. Finally, three obviously upregulated miRNAs, including miR-19a-3p, miR-17-3p, and miR-18b-5p were identified as potentially involved in the LNM of ESCC and were regarded as candidate miRNAs capable of predicting regional LNM in ESCC. The detailed data of the 19 miRNAs verified are showed in Table 3.
Table 3
| Systematic name | Group 1† (mean ± SD) | Group 2§ (mean ± SD) | Expression tendency | P value | Consistency with microarray data |
|---|---|---|---|---|---|
| miR-19a-3p | 2.7755±1.9353 | 4.3625±2.4464 | Up | 0.0142 | Consistent |
| miR-17-3p | 11.5405±7.1147 | 20.8601±11.6442 | Up | 0.0015 | Consistent |
| miR-18b-5p | 3.8370±3.5448 | 7.4825±4.7258 | Up | 0.0034 | Consistent |
| miR-4440 | 15.0672±9.7576 | 28.5884±12.8204 | Up | 0.0001 | Inconsistent |
| miR-6127 | 1.8697±2.4215 | 7.0070±10.2709 | Up | 0.0219 | Inconsistent |
| miR-18a-5p | 11.8070±17.2393 | 15.4644±12.1872 | Up | 0.3907 | Consistent |
| miR-4728-5p | 12.1957±8.2501 | 10.1191±5.7729 | Down | 0.3077 | Consistent |
| miR-301a-3p | 4.0238±5.3562 | 6.0518±4.9430 | Up | 0.1706 | Consistent |
| miR-1587 | 2.8291±4.4520 | 2.0499±3.4587 | Down | 0.4929 | Consistent |
| miR-6779-5p | 11.1996±15.6308 | 6.5792±9.1691 | Down | 0.2100 | Consistent |
| miR-1249-5p | 11.5868±25.2595 | 1.9651±2.0145 | Down | 0.0696 | Consistent |
| miR-25-3p | 5.8477±7.8516 | 5.9716±15.2436 | Up | 0.9713 | Consistent |
| miR-590-5p | 1.7158±1.9584 | 1.9663±3.1066 | Up | 0.7346 | Consistent |
| miR-106b-5p | 3.1245±3.8459 | 5.2519±5.1641 | Up | 0.1056 | Consistent |
| miR-1290 | 1.4002±1.2459 | 1.1800±1.3934 | Down | 0.5588 | Consistent |
| miR-93-5p | 2.4981±1.8751 | 4.1980±3.9970 | Up | 0.0626 | Consistent |
| miR-197-5p | 3.0791±4.4242 | 7.5768±11.3776 | Up | 0.0750 | Inconsistent |
| miR-17-5p | 3.8699±6.0198 | 3.1352±2.3148 | Down | 0.5716 | Inconsistent |
| miR-20a-5p | 2.3972±4.5983 | 1.5096±1.1455 | Down | 0.3537 | Inconsistent |
†, Group 1: expression levels of miRNAs in cancer tissues without lymph node metastasis; §, Group 2: expression levels of miRNAs in cancer tissues with lymph node metastasis.
Relations between candidate miRNAs and clinicopathologic features
To further verify the specificity of the three candidate miRNAs capable of predicting regional LNM in ESCC, the relations between candidate miRNAs and clinicopathologic features were also analyzed. The results showed that the three candidate miRNAs were not significantly associated with all other clinicopathologic features, except TNM stage, which was determined by the N stage, so the three candidate miRNAs could be regarded as tumor-specific miRNAs capable of predicting regional LNM of ESCC. All data are detailed in Table 4.
Table 4
| Main features | n | hsa-miR-19a-3p | hsa-miR-17-3p | hsa-miR-18b-5p | |||||
|---|---|---|---|---|---|---|---|---|---|
| Mean ± SD | P value | Mean ± SD | P value | Mean ± SD | P value | ||||
| Gender | 0.784 | 0.612 | 0.797 | ||||||
| Male | 40 | 3.6147±2.4346 | 15.8129±11.0116 | 5.5763±4.5577 | |||||
| Female | 10 | 3.3863±1.9273 | 17.7501±9.4024 | 5.9937±4.6209 | |||||
| Age (year) | 0.156 | 0.613 | 0.954 | ||||||
| <60 | 20 | 2.9944±2.0373 | 15.2544±10.2401 | 5.7061±4.8784 | |||||
| ≥60 | 30 | 3.9521±2.4576 | 16.8309±11.0367 | 5.6289±4.3607 | |||||
| Primary location | 0.457 | 0.93 | 0.382 | ||||||
| Lower thoracic esophagus | 26 | 3.8069±2.0788 | 16.3295±10.8015 | 6.2036±4.1978 | |||||
| Midthoracic esophagus | 24 | 3.3113±2.5869 | 16.0603±10.7074 | 5.0706±4.8773 | |||||
| Pathology grade | 0.401 | 0.593 | 0.756 | ||||||
| Moderate and well | 30 | 3.7973±2.4582 | 15.5333±10.5382 | 5.4949±3.9463 | |||||
| Poor | 20 | 3.2266±2.1258 | 17.2009±11.0041 | 5.9071±5.3804 | |||||
| Clinical subtype | 0.806 | 0.07 | 0.662 | ||||||
| Ulcerative type | 23 | 3.6581±1.9898 | 13.2506±8.8091 | 5.3519±4.0881 | |||||
| Medullary and other type | 27 | 3.4932±2.6133 | 18.7131±11.5639 | 5.9219±4.9303 | |||||
| T stage | 0.303 | 0.911 | 0.726 | ||||||
| T1–2 | 10 | 2.8846±2.0499 | 16.5407±10.3503 | 5.2051±4.4572 | |||||
| T3 | 40 | 3.7401±2.3823 | 16.1152±10.8475 | 5.7735±4.5919 | |||||
| TNM stage† | 0.008 | 0.002 | 0.002 | ||||||
| I–II | 26 | 2.7521±1.8999 | 11.7499±7.0523 | 3.8157±3.4749 | |||||
| III–IV | 24 | 4.4539±2.4551 | 21.0215±11.8661 | 7.6575±4.7441 | |||||
| Tumor size (cm2) | – | −0.047§ | 0.745 | 0.102§ | 0.479 | 0.034§ | 0.815 | ||
†, postoperative pathological stage; §, Pearson correlation coefficient.
Potential mechanism of the tumor-specific miRNAs
For the three tumor-specific miRNAs capable of predicting the regional LNM of ESCC, we could not obtain the potential target genes of miR-19a-3p and miR-18b-5p, according to the above miRNA target gene prediction criteria, but 858 potential target mRNAs were obtained for miR-17-3p. The 858 target genes of miR-17-3p were enriched in 180 KEGG pathways, which included some pathways associated with cancer such as the Wnt signaling pathway, PI3K-Akt signaling pathway, AMPK signaling pathway, and MAPK signaling pathway, etc. All these were possibly involved in the miR-17-3p potential mechanism to regulate LNM in ESCC.
Discussion
The tendency toward extensive regional LNM is an important clinical characteristic of ESCC and also a major cause of failure in surgery and radiotherapy (5,6), which results in a disappointing 5-year overall survival rate (3,4). Therefore, the search for appropriate molecular biomarkers to predict regional LNM of ESCC has become the focus at present. Li et al. reviewed potential biomarkers for predicting regional LNM in ESCC in some studies in recent years (19). However, these biomarkers are still not able to guide the treatment of regional LNM in ESCC due to the lack of pertinence and specificity on LNM status. Based on the cognition that the molecular biological properties of cancer determine the biological behaviors of cancer and the tumor specificity of miRNA in tumorigenesis, development, invasion, and metastasis (12,15,16), this microarray analysis of miRNA based on LNM status obtained three tumor-specific miRNAs for predicting regional LNM in ESCC, which would guide the selective lymphatic dissection or irradiation of ESCC, change the treatment modalities of ESCC, and be conducive to bring great progress in the treatment of ESCC.
In this study, to assure the reliabilities of the experimental results, strict quality control measures were taken for sample selection, pathological sampling, sample storage, RNA isolation, and microarray analysis to eliminate any possible interference with the experimental results. Finally, we obtained the expected results, which were intuitively presented with cluster plot, Circos plot, volcano plot, and scatter plot as shown in Figure 1.
A comparative microarray analysis identified 62 miRNAs that were differentially expressed in ESCC tissues with LNM compared to those without LNM. In combination with the target prediction analysis based on miRWalk2.0, 50 miRNAs and 8,637 target genes were considered to have potential regulatory functions in the regional LNM of ESCC. Subsequent GO and KEGG pathway analyses showed the functions of these potential target mRNAs and their enriched pathways. Several pathways were significantly involved in the pathogenesis of cancer. These results, on the one hand, exemplify the need for this experiment, and on the other hand, have provided valuable research evidence of a select number of miRNAs and their target genes for further studies.
Among the 62 miRNAs that had underlying regulatory function in the regional LNM of ESCC according to the screening criteria, 19 miRNAs, which accounted for almost one-third of differentially expressed miRNAs identified, were selected for further qRT-PCR verification. In the verification process, an additional 50 samples were used, therefore, the results of this confirmatory experiment not only verified the consistency between the qRT-PCR verification data and microarray data, but also verified the expression levels of these screened miRNAs in ESCC tissue. Finally, the results of this confirmatory experiment showed good consistency between the qRT-PCR verification data and the microarray data.
After excluding the inconsistent miRNAs between the qRT-PCR verification data and microarray data, miRNA-17-3p, miRNA-19a-3p, and miRNA-18b-5p were identified as significantly upregulated in ESCC. Further univariate analysis among the three miRNAs and clinicopathologic features showed that the three upregulated miRNAs were not significantly associated with all other clinicopathologic features except the TNM stage, which was determined by N status. These results indicated that miRNA-17-3p, miRNA-19a-3p, and miRNA-18b-5p were involved in the regulation of regional LNM of ESCC, and could be identified as tumor-specific miRNAs used for the prediction of regional LNM in ESCC.
Among the three tumor-specific miRNAs, we could not obtain the target genes of miRNA-19a-3p and miRNA-18b-5p from target prediction analysis based on miRWalk2.0. However, several studies (20-23) revealed that miRNA-19a-3p was involved in regulating the biological processes of a few benign diseases and malignant tumors via different mechanisms. Wang et al. reported that miRNA-19a-3p might contribute to the pathogenesis of preeclampsia during pregnancy via the lncRNA-PSG10P/miRNA-19a-3p/IL1RAP pathway (20). Chen et al. reported that miRNA-19a-3p promoted rheumatoid arthritis fibroblast-like synoviocytes via targeting SOCS3 (21). Bai et al. found that miRNA-19a-3p could enhance the malignancy grade of human ovarian cancer cells via inhibiting the expression of IGFBP-3 (22). Li et al. found miRNA-19a-3p could regulate cell growth and apoptosis in gastric cancer via the miRNA-19a-3p/PTEN/PI3K/AKT signaling pathway (23). Although there were many such studies, the function of miRNA-19a-3p in ESCC was not reported. With respect to miRNA-18b-5p, the number of studies is rare and only a few researchers have published reports (24-26). Zhang et al. found miR-18a-5p were significantly upregulated in colorectal cancer tissues and significantly elevated in colorectal cancer plasma exosomes, but its function and mechanism were unclear (24). Xue et al. confirmed that miRNA-18b-5p could regulate lung adenocarcinoma cell proliferation via the miR-18b-5p/VMA21 axis (25). Cochetti et al. found that miR-18b-5p owned the highest sensitivity and specificity for the prediction of prostate cancer, and decreased in expression concomitantly with an increase in malignancy, the mechanism was not studied (26). Finally, for tumor-specific miRNA-17-3p, 858 potential target mRNAs were obtained based on the present study criteria, and these target genes enriched many tumor-related pathways. Overall, this indicates that the function and regulatory mechanism of miRNA-17-3p are intricate and extensive in the pathogenesis of cancer. However, over the past ten years, studies associated with miRNA-17-3p focused more on benign diseases such as inhibiting angiogenesis by downregulating Flk-1 (27), inhibiting mouse cardiac fibroblast senescence by targeting Par4 (28), and promoting keratinocyte cell growth and metastasis in cutaneous wound healing by targeting MYOT and regulating the Notch1/NF-κB pathways (29), and so on. Moreover, only a few studies report results associated with miRNA-17-3p in cancer including prostate cancer (30), colon cancer (31), and glioblastoma (32). However, among the studies associated with miRNA-17-3p, there was no research related to ESCC. All the studies above indicated that the potential mechanisms of the three tumor-specific miRNAs regulating regional LNM of ESCC were worthy of being studied and provides evidence for further studies. Moreover, further function and mechanism studies will be conducive to confirm the application value of the three tumor-specific miRNAs and provide theoretical foundation for their clinical application.
Conclusions
In summary, the present microarray analysis of miRNAs based on the LNM status of ESCC identified three tumor-specific miRNAs for the prediction of regional LNM in ESCC and provides significant evidence for future studies of the potential mechanism, which warrants further investigation.
Acknowledgments
We thank International Science Editing (http://www.internationalscienceediting.com) for editing this manuscript.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-1969
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-1969
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-1969
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-1969). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committees of the Shandong Cancer Hospital and the Second Hospital of Shandong University [registration number: KYLL-2017(GJ)P-0036] and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359-86. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Pennathur A, Gibson MK, Jobe BA, et al. Oesophageal carcinoma. Lancet 2013;381:400-12. [Crossref] [PubMed]
- Rustgi A, El-Serag HB. Esophageal carcinoma. New Engl J Med 2015;372:1472-3. [PubMed]
- Wang Y, Zhu L, Xia W, et al. Anatomy of lymphatic drainage of the esophagus and lymph node metastasis of thoracic esophageal cancer. Cancer Manag Res 2018;10:6295-303. [Crossref] [PubMed]
- Tachimori Y. Pattern of lymph node metastases of squamous cell esophageal cancer based on the anatomical lymphatic drainage system: efficacy of lymph node dissection according to tumor location. J Thorac Dis 2017;9:S724-30. [Crossref] [PubMed]
- Ma GW, Situ DR, Ma QL, et al. Three-field vs two-field lymph node dissection for esophageal cancer: a meta-analysis. World J Gastroenterol 2014;20:18022-30. [Crossref] [PubMed]
- Ji X, Cai J, Chen Y, et al. Lymphatic spreading and lymphadenectomy for esophageal carcinoma. World J Gastrointest Surg 2016;8:90-4. [Crossref] [PubMed]
- Van De Voorde L, Larue RT, Pijls M, et al. A qualitative synthesis of the evidence behind elective lymph node irradiation in oesophageal cancer. Radiother Oncol 2014;113:166-74. [Crossref] [PubMed]
- Gockel I, Sgourakis G, Lyros O, et al. Dissection of lymph node metastases in esophageal cancer. Expert Rev Anticancer Ther 2011;11:571-8. [Crossref] [PubMed]
- Kayani B, Zacharakis E, Ahmed K, et al. Lymph node metastases and prognosis in oesophageal carcinoma--a systematic review. Eur J Surg Oncol 2011;37:747-53. [Crossref] [PubMed]
- Denlinger CE, Thompson RK. Molecular basis of esophageal cancer development and progression. Surg Clin North Am 2012;92:1089-103. [Crossref] [PubMed]
- Conrad R, Barrier M, Ford LP. Role of miRNA and miRNA processing factors in development and disease. Birth Defects Res C Embryo Today 2006;78:107-17. [Crossref] [PubMed]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006;94:776-80. [Crossref] [PubMed]
- Lopez-Camarillo C, Marchat LA, Arechaga-Ocampo E, et al. MetastamiRs: non-coding miRNAs driving cancer invasion and metastasis. Int J Mol Sci 2012;13:1347-79. [Crossref] [PubMed]
- Chan SH, Wang LH. Regulation of cancer metastasis by microRNAs. J Biomed Sci 2015;22:9. [Crossref] [PubMed]
- Mei LL, Qiu YT, Zhang B, et al. MicroRNAs in esophageal squamous cell carcinoma: Potential biomarkers and therapeutic targets. Cancer Biomark 2017;19:1-9. [Crossref] [PubMed]
- Dias F, Morais M, Teixeira AL, et al. Involving the microRNA targetome in esophageal cancer development and behavior. Cancers (Basel) 2018;10:381. [Crossref] [PubMed]
- Li J, Qi Z, Hu YP, et al. Possible biomarkers for predicting lymph node metastasis of esophageal squamous cell carcinoma: a review. J Int Med Res 2019;47:544-56. [Crossref] [PubMed]
- Wang N, Li R, Xue M. Potential regulatory network in the PSG10P/miR-19a-3p/IL1RAP pathway is possibly involved in preeclampsia pathogenesis. J Cell Mol Med 2019;23:852-64. [Crossref] [PubMed]
- Chen Y, Wang W, Chen Y, et al. MicroRNA-19a-3p promotes rheumatoid arthritis fibroblast-like synoviocytes via targeting SOCS3. J Cell Biochem 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Bai R, Cui Z, Ma Y, et al. The NF-κB-modulated miR‐19a‐3p enhances malignancy of human ovarian cancer cells through inhibition of IGFBP‐3 expression. Mol Carcinog 2019;58:2254-65. [Crossref] [PubMed]
- Li X, Yan X, Wang F, et al. Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. J Cell Mol Med 2019;23:2920-32. [Crossref] [PubMed]
- Zhang H, Zhu M, Shan X, et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019;687:246-54. [Crossref] [PubMed]
- Xue M, Tao W, Yu S, et al. lncRNA ZFPM2-AS1 promotes proliferation via miR-18b-5p/VMA21 axis in lung adenocarcinoma. J Cell Biochem 2020;121:313-21. [Crossref] [PubMed]
- Cochetti G, Poli G, Guelfi G, et al. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther 2016;9:7545-53. [Crossref] [PubMed]
- Yin R, Wang R, Guo L, et al. MiR-17-3p inhibits angiogenesis by downregulating Flk-1 in the cell growth signal pathway. J Vasc Res 2013;50:157-66. [Crossref] [PubMed]
- Du WW, Li X, Li T, et al. The microRNA miR-17-3p inhibits mouse cardiac fibroblast senescence by targeting Par4. J Cell Sci 2015;128:293-304. [Crossref] [PubMed]
- Yan H, Song K, Zhang G. MicroRNA-17-3p promotes keratinocyte cells growth and metastasis via targeting MYOT and regulating Notch1_NF-κB pathways. Pharmazie 2017;72:543-9. [PubMed]
- Yang X, Du WW, Li H, et al. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res 2013;41:9688-704. [Crossref] [PubMed]
- Lu D, Tang L, Zhuang Y, et al. MiR-17-3p regulates the proliferation and survival of colon cancer cells by targeting Par4. Mol Med Rep 2018;17:618-23. [PubMed]
- Delen E, Doganlar O, Doganlar ZB, et al. Inhibition of the invasion of human glioblastoma U87 cell line by Ruxolitinib: a molecular player of miR-17 and miR-20a regulating JAK/STAT pathway. Turk Neurosurg 2020;30:182-9. [PubMed]


