Gene network screening of bladder cancer via modular analysis
Introduction
Bladder cancer (BC) is one of the most common cancers of the urinary system. BC is the 13th most common cause of cancer-related death worldwide (1). The majority of BC is non-muscle invasive bladder cancers (NMIBC), which tend to have a high rate of recurrence after primary tumor resection. The 5-year overall survival (OS) for NMIBC patients is nearly 90%, and 60% to 70% for muscle invasive bladder cancers (MIBC) patients (2). Approximately 50% of patients occurred distant metastasis after radical cystectomy (3). For decades, the outcome or treatment for BC has not progressed much (4). Cisplatin was tested in neoadjuvant chemotherapy for MIBC since 1980s, and still used after cystectomy or metastatic patients as the first line option (5). The use of platinum-based chemotherapy has been limited because of neutropenia, peripheral neuropathy and mucositis (5). The progression, metastasis and drug resistance also barricade the treatment of BC.
Therefore, more accurate tumor characterization and stratification of BC patients for selection of more appropriate treatments are required. BC is a very heterogeneous disease due to clinical history, the pathological features and the molecular mechanisms involved in each case differ (6). FGFR is an oncogene and play important roles in cell proliferation, migration and invasion (7). FGFR3 mutations are highly associated with low-grade non-muscle invasive urothelial carcinoma (8). Medicines such as FGFR1 and FGFR3 inhibitors have been developed to treat BC, but these drugs are still in the continuation phase of clinical trials (9,10).
Gene amplifications have been found in EGFR and MET (11). miRNAs are long non-coding RNA gene products which can serve as oncogenes or tumor suppressors, it regulates target genes by binding to specific sites.
An increasing number of studies have implied that miRNAs might be the potential biomarkers and molecular therapeutic targets for BC. Gene pairs such as EGFR and c-MET are regulated by microRNA-23b/27b which contribute to BC oncogenesis and metastasis (12).
Numerous genes and miRNAs are involved in the occurrence and development of BC, the complicated regulatory mechanism remains unclear. Previous study has constructed a protein-protein interaction network by differentially expressed genes of BC. PCNA, TOP2A, CCND1 and CDH1 were found to be hub genes in the network (13).
Although much has been known about single gene or miRNA in BC, much less is on the roles of paired significant genes and miRNAs. In this study, we utilized genetic associated genes network construction to identify the gene correlation and OS time in BC, and analyzed the miRNAs which might regulate the significant modules and hub genes. Multi-level molecular mechanism was also explored.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-2822).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Gene collection and network construction
The data for this study are from the National Center for Biotechnology Information (NCBI)’s Online Mendelian Inheritance in Man (OMIM) database http://www.ncbi.nlm.nih.gov/omim), which is a knowledge database of human genes and genetic disorders. Disease-associated genes were performed via multiple text-based Searching in Agilent Literature Search software version 3.2.2 (http://www.agilent.com/labs/research/litsearch.html), by which the gene network was constructed.
Module division
MCODE (http://baderlab.org/Software/MCODE) version 1.32 was used for computational analysis of network for the gene complex detection. Genes with common biological processes or pathways were divided into the same module.
Functional enrichment
DAVID (http://david.abcc.ncifcrf.gov) was used for Gene ontology (GO) and pathway analysis (14). Parameters: Count, 2; EASE, 0.01; and species and background, Homo sapiens. The biological processes and pathways were ranked by P values.
Overall survive time and correlation analysis of hub genes
The OS time of hub gene expression was analyzed by GEPIA (15). Parameters: hazards ratio (HR): yes, 95% CI: yes, axis units: month. The study used Pearson Correlation Coefficient for correlated calculation of the hub genes in the same module (Bladder Urothelial Carcinoma samples compared with normal samples).
miRNA prediction
We enriched the modules and predict the regulated miRNAs by Cluepedia (edge score =0.6, threshold =3). Interactions within each pathway can be investigated and new potential associations are revealed through gene/miRNA enrichments (16).
Statistical analysis
The study used Pearson Correlation Coefficient for correlated calculation of the hub genes in the same module (Bladder Urothelial Carcinoma samples compared with normal samples). The correlation of hub genes was used by non-log scale, and log-scale axis for visualization. Mantel–Cox test was used for the hypothesis evaluation of OS analysis.
Results
General gene information
A total of 187 bladder cancer-associated genes (including NMIBC and MIBC) were got from OMIM (Appendix 1). And 177 of which link to homologene based on a common GeneID, 23 genes link to UniSTS which based on markers cited in the OMIM record, 56 genes link to variation data in dbSNP (https://www.ncbi.nlm.nih.gov/snp). UniSTS is a large STS database comprised of both GenBank STS sequence entries and published STS maps (17). dbSNP contains human single nucleotide variations, microsatellites, and small-scale insertions for both common variations and clinical mutations (https://www.ncbi.nlm.nih.gov/snp).
BC gene network
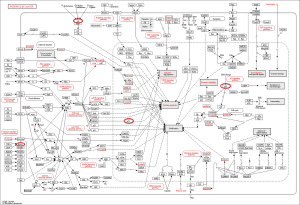
Inputting 187 BC-associated genes into the Agilent Literature Search, the BC gene network contains 1,289 nodes and 7,164 edges (Figure 1). The average number of node neighbors is 10.438, and the isolated nodes number is 76.

Module analysis
Dense regions of the BC gene network were divided by MCODE. Totally, 35 modules found in the network (Appendix 2). Three modules (modules 1, 2 and 3) have the largest nodes were detected (Figure 2). EGFR, AR, MET, RELA, TP53 and TSG101 are hub genes (edges above 10) of the largest 3 modules.

Enrichment analysis
A total of 216 functional annotations and 95 pathways were found in the enrichment analysis of the most significant top 3 modules (https://cdn.amegroups.cn/static/public/TCR-20-2822-1.pdf). Negative regulation of apoptotic process and pathways in cancer are the most significant biological process and pathway separately (Figure 3). The hub genes in the top 3 modules involved in the significant processes such as regulation of cell cycle and positive regulation of transcription from RNA polymerase II promoter (Table 1).

Table 1
| TP53-TSG101 | AR-RELA | MET-EGFR |
|---|---|---|
| GO: 0051726 regulation of cell cycle (1.08E-14) | GO: 0045944 positive regulation of transcription from RNA polymerase II promoter (7.56E-10) | GO: 0045944 positive regulation of transcription from RNA polymerase II promoter (7.56E-10) |
| GO: 0051222 positive regulation of protein transport (5.27E-7) | GO: 0008284 positive regulation of cell proliferation (1.22E-5) | GO: 0048729 tissue morphogenesis (4.60E-4) |
| GO: 0005730 positive regulation of protein transport and nucleolus (8.40E-3) | GO: 0051092 positive regulation of NF-κB transcription factor activity (1.40E-2) | GO: 0044255 cellular lipid metabolic process (5.50E-3) |
GO, gene ontology.
OS time and correlation to hub genes
The results demonstrate that BC patients with low-expressed TSG101 have longer OS, and are associated with TP53. Low-expressed RELA and over-expressed AR patients have a higher survival time. Low-expressed TSG101 patients have a longer survival time (Figure 4).
miRNA prediction
miRNA17, miRNA20a and miRNA15a were found to regulate module 1, miRNA15a and has-let-7b regulating module 2, miRNA15a and miRNA16 regulating module 3 (Figure 5).

Discussion
By the investigation of human genome-wide functional microarray or RNA-seq gene expression in pathway databases, TP53, AR and RELA were found as transcriptional targets (18). In the present study, the hub genes, miRNAs and pathways associated with BC were identified.
TP53 and TSG101 are hub genes of module 1, there is a positive correlation of gene expression between them. TP53 is involved in the regulation of cell cycle and apoptosis. The expressions of TP53 in NMIBC cells (KK47 and RT4) were lower than those in MIBC cells (T24, 5637, and UM-UC-3) (19). Overexpression of TP53 is related to poor survival in patients with advanced BC (20). Mutations in the TP53 have been observed more frequently in invasive high grade BC compared with low grade BC (21). TSG101 is a common target of splicing defects, the stress-activated TP53 can regulate TSG101 splicing process (22). Meanwhile, TSG101 attenuates p53 signaling (23), and the TSG101 transcripts is correlated with tumor grade and p53 mutation in breast cancer (24). The GO analysis for The TOP 3 modules demonstrated that TP53 and TSG101 are involved in the processes including regulation of cell cycle, positive regulation of protein transport and nucleolus.
AR and RELA are hub genes of module 2. AR is a nuclear steroid hormone receptor and play key roles in the occurrence and progression of many cancers (25). Systems biology modeling demonstrated that RELA and AR are hub genes of the radiation-specific biomarkers and related to radio-sensitization drugs (26). Interleukin-1 (IL-1) is implicated in prostate cancer initiation and progression, RELA can regulate IL-1-mediated AR repression in prostate cancer cells (27). Meanwhile, AR declined the angiogenic potential of cancer cells. The activation of AR decreases the expression of RELA, and reduced its transcriptional activity which is an anti-tumor mechanism (28). AR together with RELA involved in 5 biological processes, the most significant is positive regulation of transcription from RNA polymerase II promoter, which is equal to MET and EGFR.
MET and EGFR are hub genes of module 3. MET is associated with the progression, treatment effect and OS of cancers. Urinary soluble MET level of BC patients is higher than patients without BC (29). EGFR and c-Met signaling pathways can be regulated by miR-23b/27b, the decreased expression of which may enhance cancer cell proliferation and migration (12). MET together with EGFR involve in 3 biological processes, including positive regulation of transcription from RNA polymerase II promoter, tissue morphogenesis, cellular lipid metabolic process.
Previous study confirmed that miRNAs can be critical players in the prognosis and diagnosis of BC (30). miRNA16 inhibited the proliferation, migration, and invasion of CRC cells by downregulating ITGA2 (31).
MiRNA-17, miR-20a, miR-15a, let-7b, miR-16 were predicted regulating the top 3 modules of BC. mir-17 and mir-15a were found significantly correlated with the OS of BC (32).
Pathways in cancer (hsa05200) is the most significant pathway according to P value. Hub genes, such as AR, TP53 and EGFR, were found to take part in the apoptosis and cytokine-cytokine receptor interaction of the pathway.
In conclusion, our study revealed multiple possible significant functional mechanisms in the BC development. The combined pattern of hub genes, miRNAs, significant processes and pathways supply new drug targets and treatments for further study.
Acknowledgments
We sincerely appreciate Dr. Hanze Zhang for his valuable language editing for this manuscript.
Funding: This study was supported by grants from
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2822
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2822). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Magers MJ, Lopez-Beltran A, Montironi R, et al. Staging of bladder cancer. Histopathology 2019;74:112-34. [Crossref] [PubMed]
- Yafi FA, Aprikian AG, Chin JL, et al. Contemporary outcomes of 2287 patients with bladder cancer who were treated with radical cystectomy: a Canadian multicentre experience. BJU Int 2011;108:539-45. [Crossref] [PubMed]
- Ebrahimi H, Amini E, Pishgar F, et al. Global, Regional and National Burden of Bladder Cancer, 1990 to 2016: Results from the GBD Study 2016. J Urol 2019;201:893-901. [Crossref] [PubMed]
- Flaig TW, Spiess PE, Agarwal N, et al. Bladder Cancer, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2020;18:329-54. [Crossref] [PubMed]
- Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nature Reviews Cancer 2015;15:25-41. [Crossref] [PubMed]
- Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116-29. [Crossref] [PubMed]
- Tomlinson DC, Baldo O, Harnden P, et al. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J Pathol 2007;213:91-8. [Crossref] [PubMed]
- Krook MA, Lenyo A, Wilberding M, et al. Efficacy of FGFR Inhibitors and Combination Therapies for Acquired Re-sistance in FGFR2-Fusion Cholangiocarcinoma. Mol Cancer Ther 2020;19:847-57. [Crossref] [PubMed]
- Nogova L, Sequist LV, Perez Garcia JM, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 Kinase Inhibitor, in Patients With Advanced Solid Tumors Harboring Genetic Alterations in Fibroblast Growth Factor Receptors: Results of a Global Phase I, Dose-Escalation and Dose-Expansion Study. J Clin Oncol 2017;35:157-65. [Crossref] [PubMed]
- Reis H, van der Vos KE, Niedworok C, et al. Pathogenic and targetable genetic alterations in 70 urachal adeno-carcinomas. Int J Cancer 2018;143:1764-73. [Crossref] [PubMed]
- Chiyomaru T, Seki N, Inoguchi S, et al. Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int J Oncol 2015;46:487-96. [Crossref] [PubMed]
- Tang F, He Z, Lei H, et al. Identification of differentially expressed genes and biological pathways in bladder cancer. Mol Med Rep 2018;17:6425-34. [Crossref] [PubMed]
- Huang L, Xie D, Yu Y, et al. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res 2018;46:D1117-d20. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 2013;29:661-3. [Crossref] [PubMed]
- Rotmistrovsky K, Jang W, Schuler GD. A web server for performing electronic PCR. Nucleic Acids Res 2004;32:W108-12. [Crossref] [PubMed]
- Shmelkov E, Tang Z, Aifantis I, et al. Assessing quality and completeness of human transcriptional regulatory pathways on a genome-wide scale. Biol Direct 2011;6:15. [Crossref] [PubMed]
- Yan L, Li Q, Yang J, et al. TPX2-p53-GLIPR1 regulatory circuitry in cell proliferation, invasion, and tumor growth of bladder cancer. J Cell Biochem 2018;119:1791-803. [Crossref] [PubMed]
- Lorenzo-Romero JG, Salinas-Sánchez AS, Giménez-Bachs JM, et al. Prognostic implications of p53 gene mutations in bladder tumors. J Urol 2003;169:492-9. [Crossref] [PubMed]
- Proctor I, Stoeber K, Williams GH. Biomarkers in bladder cancer. Histopathology 2010;57:1-13. [Crossref] [PubMed]
- Moyret-Lalle C, Duriez C, Van Kerckhove J, et al. p53 induction prevents accumulation of aberrant transcripts in cancer cells. Cancer Res 2001;61:486-8. [PubMed]
- Gray TA, Alsamman K, Murray E, et al. Engineering a synthetic cell panel to identify signalling components re-programmed by the cell growth regulator anterior gradient-2. Mol Biosyst 2014;10:1409-25. [Crossref] [PubMed]
- Turpin E, Dalle B, de Roquancourt A, et al. Stress-induced aberrant splicing of TSG101: association to high tumor grade and p53 status in breast cancers. Oncogene 1999;18:7834-7. [Crossref] [PubMed]
- Chang C, Lee SO, Yeh S, et al. Androgen receptor (AR) differential roles in hormone-related tumors including prostate, bladder, kidney, lung, breast and liver. Oncogene 2014;33:3225-34. [Crossref] [PubMed]
- Eschrich S, Zhang H, Zhao H, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys 2009;75:497-505. [Crossref] [PubMed]
- Thomas-Jardin SE, Dahl H, Kanchwala MS, et al. RELA is sufficient to mediate interleukin-1 repression of androgen receptor expression and activity in an LNCaP disease progression model. Prostate 2020;80:133-45. [Crossref] [PubMed]
- Nelius T, Filleur S, Yemelyanov A, et al. Androgen receptor targets NFkappaB and TSP1 to suppress prostate tumor growth in vivo. Int J Cancer 2007;121:999-1008. [Crossref] [PubMed]
- Matsumoto K, Umitsu M, De Silva DM, et al. Hepatocyte growth factor/MET in cancer progression and biomarker discovery. Cancer Sci 2017;108:296-307. [Crossref] [PubMed]
- Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci 2019;110:408-19. [Crossref] [PubMed]
- Xu Y, Shen L, Li F, et al. microRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. 2019;234:21380-94.
- Yin XH, Jin YH, Cao Y, et al. Development of a 21-miRNA Signature Associated With the Prognosis of Patients With Bladder Cancer. Front Oncol 2019;9:729. [Crossref] [PubMed]