
Effect of hematopoietic progenitor cells on the biological characteristics of colon cancer tumor stem cells
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors in the world, and liver metastasis is the most common form of CRC metastasis (1). Studies have shown that liver metastasis can occur in ~20% of newly diagnosed CRC patients within half a year (2,3). In addition, >50% of patients with advanced CRC have a median survival of only 21.3 months (4). Currently, the “seed and soil” theory proposed by British surgeon Dr. Stephen Paget is generally accepted as the underlying developmental mechanism of colorectal cancer liver metastases (CRLM) (5). On this basis, the “cancer stem cell theory” has been expanded in recent years (6). Cancer stem cells (CSCs) are a group of tumor cells that are self-sustaining and possess the ability to differentiate into different types of cells. CSCs are the source of the initiation and maintenance of tumor growth and serve a decisive role in tumor recurrence and metastasis (7).
CSC function may be induced by specific microenvironmental signals and cellular interactions in the tumor niche (8). CSCs were considered as “seed”, meanwhile the microenvironment of the target organ were “soil”. Based on this theory, it was shown that primary tumor cells secrete specific cytokines prior to metastasis to mobilize a group of myeloid precursor cells into the circulatory system, which are then recruited into the tissues of the target organ to remodel the premetastatic site microenvironment. This group of cells serve an important role in the formation of a microenvironment suitable for the growth of tumor cells in the premetastatic target organs (9,10). Kaplan et al. identified this group of hematopoietic progenitor cells (HPCs) by using mouse animal models that express molecular markers, such as VEGFR1, CD133, CD11b and scal-1 (11). It was reported that HPCs could facilitate colorectal cancer procession (12). However, few studies explore the contribution of colon cancer stem cells (CCSCs) on this phenomenon and the underlying mechanism is still undefined.
The aim of the present study was to investigate the effect of HPCs on the biological behavior of CCSCs and to examine the molecular mechanisms of metastasis in CCSCs. The results may assist in the identification of novel molecular targets for the treatment of metastatic colorectal cancer. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2810).
Methods
Cell lines and reagents
The human colon cancer cell line HCT116, was gifted by the Institute of Oncology, Fujian Provincial Cancer Hospital. HPCs and monocytes cryopreservation solution was obtained from iXCells Biotechnologies, Inc. The CD133+ HPC population was verified by flow cytometry. Human basic fibroblast growth factor (bFGF) and human epidermal growth factor (EGF) were obtained from PeproTech, Inc. B-27 supplement (50×) was obtained from Gibco; Thermo Fisher Scientific, Inc. Anti-human CD133-PE were purchased from (R&D Systems, Inc.); anti-human CD24-fluorescein isothiocyanate (FITC) was obtained from Becton, Dickinson and Company; and fluorescent secondary antibody was obtained from OriGene Technologies, Inc. Cell Counting Kit-8 (CCK-8) was obtained from Dojindo Molecular Technologies, Inc. Transwell chambers were obtained from EMD Millipore. HiFi-mmlv cDNA kit and Ultra SYBR mixture were purchased from KW-BIO. The bicinchoninic protein assay (BCA) kit was obtained from Beyotime Institute of Biotechnology. Roswell Park Memorial Institute (RPMI) 1640 medium were purchased from Hyclone, Inc. Goat serum sealant (ZLI-9022) were purchased from ZSGB-BIO, Inc. ACCUTASE cell digestive juice were purchased from Yeasen Biotech Co., Ltd., Shanghai.
CCSC culture and detection
The HCT116 cell line was subcultured in RPMI 1640 medium containing 10% FBS; cells were obtained during the logarithmic growth phase and inoculated in serum-free medium (SFM) supplemented with 10 ng/mL bFGF, 20 ng/mL EGF and 2% B27 in Gibco Dulbecco’s Modified Eagle Medium: F-12 (DMEM/F-12) (12). In SFM, the differentiated cells in HCT116 cells were apoptotic because they could not stick to the wall for growth. However, the subpopulations of CSCs with stem cell potential could survive sustainably and grow in stem cell globulin suspension (13,14). bFGF, EGF and B27 growth factors were added to SFM at specific concentrations, in which EGF and bFGF could significantly promote the proliferation of CCSCs (15). B27 can replace serum as the basic nutrient needed to stimulate cell growth without promoting differentiation of CCSCs. Previous literature has also reported that serum-free culture and separation method is suitable for CCSCs separation (16,17). HCT116 colon cancer cells were suspended in SFM as single cells and allowed to proliferate in the form of a “bud”. After 3–7 days, they gradually formed oblate-like cell spheres with a dense structure and regular edges in microscope. After 10 days, we obtained the CCSCs spheres. After three generations of continuous passages at a density of 1×104/mL, tumor cell spheres in a healthy growth state were taken for subsequent experiments at a density of 1×106/mL. Mouse anti-human monoclonal antibody, CD133-phycoerythrin (PE) was added to the experimental group, and IgG was used to treat the control group. The two groups were incubated at room temperature in the dark for 60 min. After washing with 3% FBS-PBS twice, the number of CD133+ cells was analyzed by flow cytometry (FC500, Beckman, Inc., Fullerton, CA, USA). The number of CD24+ cells in both groups were also assessed using the same method with the CD24 specific antibody.
Immunofluorescence
The HCT116 cell spheres were grown for 7–10 days, and were then cultured using the hanging drop method in normal goat serum sealant and sealed at room temperature for 30 min. The primary anti-CD133 antibody was added to each well at 1:50, incubated overnight at 4 °C in the dark, and subsequently washed twice with PBS buffer. The secondary fluorescent antibody was added to each well at 1:100, incubated in dark for 60 min at room temperature. The intensity of red fluorescence after CD133-PE labeling was observed under a fluorescence microscope. The fluorescence intensity of CD24 antibody markers in tumor cells was detected using the same method except with an anti-CD24 primary antibody instead.
CCK-8 assay
A single-cell suspension of the HCT116 cell spheres were prepared, adjusted to a cell density of 5×103/mL, 200 μL per well, plated in a 96-well plate for 12–24 h and cultured. In the experimental group, CD133+ HPCs were added 50 cells to each well, and the same number of tumor cells were added in the control group. In the blank control group, an equivalent volume of PBS was added. Subsequently, cells were cultured for 48 h, and 20 μL CCK-8 reagent was added to each well. After 2 h of culturing in a 37 °C incubator, the absorbance value of each well was measured at 450 nm using a microplate reader (Varioskan LUX, Thermo Fisher Scientific, Inc.).
Colony formation assays
Single-cell suspensions of CCSCs and CD133+ HPCs were cocultured in vitro for 48 h at a ratio of 20:1, and the control CCSCs were established. Subsequently, 400 cells per well were inoculated into 6-well plates and cultured for 10–14 days. When the colonies were visible, crystal violet dye was immediately used for staining for 15 min at room temperature. Cells were air dried, and the number of colonies formed were counted by eye (were ≥50 cells constituted a colony).
Transwell migration and invasion assays
Single-cell suspensions of the CCSC and CD133+ HPC coculture as well as the control CCSCs were resuspended in 0.1% BSA, and the cell density was adjusted to 1.5×105 cells/mL. In the Transwell chambers (8.0 μm aperture), 100 μL cell suspension was added to the upper chamber, and 600 μL RPMI 1640 supplemented with 20% FBS was added to the lower compartment. After 48 h, the cells were taken out, stained using crystal violet dye for 30 min at room temperature, and subsequently dried at room temperature. The number of cells in five randomly selected fields of view were counted using an inverted high-power microscope (200×) (TE200, NIKON), and the mean values were calculated. To assess invasion, a similar procedure was performed, except the Transwell chambers were first coated with Matrigel (BD Biosciences).
Reverse transcription-quantitative (RT-qPCR)
RT-qPCR was used to detect the mRNA expression levels of MMP-9 and VEGF in the two groups of tumor cells as indicators of invasion and metastasis. TRIpure reagent (Roche) was used to extract total RNA. The first strand cDNA from 1 µg of total RNA was synthesized using HiFi-mmlv cDNA kit which was carried out at 42 °C for 40 min and 85 °C for 5 min. Real-time fluorescent quantitative PCR was used to detect the quantity of RNA, and the results were analyzed using the 2−ΔΔCt method (18): ΔCt value is the difference between the Ct value of GAPDH gene and the Ct value of the target gene [ΔCt = Ct (target gene) − Ct (GAPDH)]. The ΔΔCt values is the difference in the Ct value between the experimental group and the control group [ΔΔCt = ΔCt (experimental group) − ΔCt (control group)].
A two-step RT-PCR kit was used for RT-PCR, and PCR amplification. PCR amplification of MMP-9 and VEGF was performed according to the protocol of the RT-PCR kit. The relative expression of genes was normalized to GAPDH. The primer sequences were shown in Table 1.
Table 1
| Gene | Primer |
|---|---|
| MMP-9 | Forward: 5'-CTGAAGGCCATGCGAACCCCA-3' |
| Reverse: 5'-GCAAAGGCGTCGTCAATCACC-3' | |
| VEGF | Forward: 5'-ACTTCTGGGCTGTTCTCGCTT-3' |
| Reverse: 5'-GCCGCCTCACCCGTCCATG-3' | |
| GAPDH | Forward: 5'-TGGTATCGTGGAAGGACT-3' |
| Reverse: 5'-AGGGATGATGTTCTGGAGA-3' |
The thermocycling conditions for PCR were as follows: predenaturation at 95 °C for 10 min; followed by 40 cycles of 15 s at 95 °C, 20 s at 60 °C and 25 s at 72 °C; with a final extension step of 72 °C for 5 min. Temperature was then ramped from 65 to 95 °C to plot the melting curve. The differences in the mRNA expression levels of MMP-9 and VEGF genes in the two groups of tumor cells were observed and evaluated.
Western blot analysis
Western blotting analysis was used to detect the protein expression levels of MMP-9, VEGF, E-cadherin and β-catenin in the two groups of tumor cells related to tumor invasion and metastasis. The two groups of cells were harvested by scraping the cells in ice-cold PBS and using RIPA lysis buffer to lyse the cells. The protein concentration was measured using a BCA kit. The lysed protein sample was loaded on an SDS-gel composed of a 10% resolving SDS-gel and a 5% loading SDS-gel. The proteins were resolved by electrophoresis at 80 V until the loading buffer had entered the resolving gel, and then the gel was run at 100 V until the bromophenol blue reached the bottom of the gel. Subsequently, the resolved proteins were transferred to a PVDF membrane which had been activated in methanol. After transfer, the membrane was cut according to the marker and location of the target band. The membranes were blocked for 1 h in Tris Buffer Saline Tween containing 5% milk powder. The membranes were then incubated overnight at 4 °C with the primary antibody (1:1,000). The membranes were washed 3 times with Tris Buffer Saline Tween and incubated with the secondary antibody (1:500) for 1h at room temperature. The membrane was allowed to dry, and signals were visualized using enhanced chemiluminescence.
Statistical analysis
All experiments were repeated at least three times and the data are presented as the mean ± the standard deviation. Datasets with only two groups were analyzed using an independent sample t-test. SPSS version 24.0 (IBM, Corp.) was used for data analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
CD133+ and CD24+ cells are significantly increased in the CCSC subsets
HCT116 colon cancer cells were suspended in SFM as single cells and allowed to proliferate in the form of a “bud”. After 3–7 days, they gradually formed oblate-like cell spheres with a dense structure and regular edges (Figure 1A). After 7–10 days, cells were digested and passed by ACCUTASE cell digestive juice in preparation for the CCSC subsets. The results of the flow cytometry showed that the counts of the CD133+ and CD24+ cells were significantly higher compared with the monolayer of cells in serum supplemented medium (SSM) (P=0.000; P=0.001, P<0.05; Figure 1B).
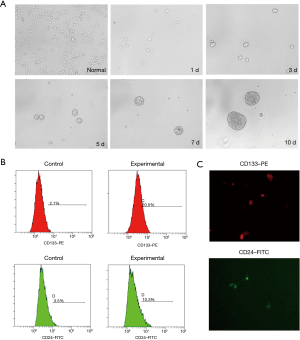
Immunofluorescence analysis showed that CD133-PE- and CD24-FITC-labeled fluorescence was visible on the surface of SFM cultured cells (Figure 1C).
Proliferation of CCSC subsets is promoted by HPCs
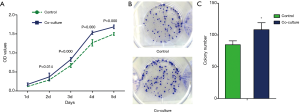
The proliferation of the two groups of cells was assessed using a CCK-8 assay at five different time points, and a growth curve was plotted. The results showed that the proliferation of CCSC subsets in the experimental group was significantly higher at the different time points (day 1 to day 5) compared with the control group (P<0.05) (Figure 2A).
Colony formation in the CCSC subsets is enhanced by HPCs
In both groups, the CCSC subsets of tumor cells formed colonies in the 6-well plates, and colony formation in the CCSC subsets in the experimental group was significantly higher than that of the control group (P=0.031) (Figure 2B,C).
Migration and invasion of CCSC subsets is enhanced following coculture with HPCs
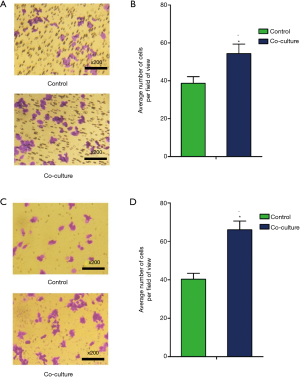
In both groups, the CCSC subsets passed through the polycarbonate membrane in the Transwell chamber, and the number of CCSC subsets passing through the polycarbonate membrane in the coculture group was significantly higher than the monoculture group (P=0.011) (Figure 3A,B). Both groups of tumor cells were able to penetrate the polycarbonate membrane and Matrigel. The number of CCSC subsets passing through the polycarbonate membrane and Matrigel in the coculture group was significantly higher compared with the monoculture group (P=0.001) (Figure 3C,D).
MMP-9, VEGF, E-cadherin and β-catenin expression in CCSC subsets is significantly increased after coculture with HPCs
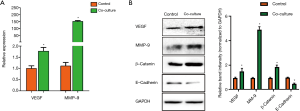
The qPCR results showed that there was a significant increase in the mRNA expression levels of MMP-9 and VEGF in the CCSC cocultured cells compared with the monoculture (MMP-9, P=0.000; VEGF, P=0.005) (Figure 4A). After coculture with HPCs, the protein expression levels of MMP-9, VEGF, E-cadherin and β-catenin in the CCSC subgroup cells of the experimental group were detected by Western blotting. The results showed that the protein expression levels of MMP-9, VEGF and β-catenin in the CCSC subgroup cells of the experimental group was significantly higher compared with the control group (MMP-9, P=0.000; VEGF, P=0.005; β-catenin, P=0.000) (Figure 4B). The results showed that the protein expression levels of E-cadherin in the CCSC subgroup cells of the experimental group was significantly lower compared with the control group (E-cadherin P=0.002) (Figure 4B).
Discussion
As early as 1998, Luzzi et al. showed that only ~0.02% of tumor cells “escaping” from the primary site could successfully reach the premetastatic target organ site (19); this shows that distant metastasis of tumor cells is extremely inefficient. However, in 2005, VEGFR1+ HPCs were observed forming clusters in specific target organs, and that they created the contours which eventually developed into metastatic sites by forming the premetastatic niche where the tumor cells in the circulatory system could settle, survive, divide and proliferate (11). Therefore, tumor stem cells and HPCs serve an important role in the formation of tumor metastasis. Successive studies have confirmed the existence of this phenomenon in the metastasis of multiple types of tumor, such as colorectal cancer, lung cancer and esophageal cancer, amongst others (20,21). With regards to distant metastasis of colorectal cancer, the liver is the most common site of distant metastasis, which is observed in about 15–25% of CRC patients at the first diagnosis (22).
Do HPCs promote liver metastasis of colon cancer? How do HPCs affect the biological behavior of CCSCs? To address these questions, CCK-8 and colony formation assays were used, and the results showed that HPCs significantly promoted the proliferation and colony formation capacity of CCSC subsets in vitro at different time periods when cocultured. When tumor metastases reach the premetastatic site to form tumor micrometastases, the tumor cells are required to invade through the basement membrane of blood vessels, which is dependent on chemotaxis. In the present study, a Transwell assay was used to assess the migratory and invasive capacity of CCSC subsets in the coculture environment, and the results showed that both migration and invasion was significantly enhanced compared with the tumor cells cultured as a monoculture. Therefore, it is hypothesized that HPCs may enhance the ability of CCSC subsets to invade the basement membrane by secreting certain cytokines, thus promoting tumor metastasis. Zhang et al. also showed the presence of HPC aggregates in colorectal cancer, and demonstrated the enhancing effect of CD133+ HPCs on tumor metastasis (12).
The degradation of the extracellular matrix is a key process required for tumor cell metastasis (23). MMP-9 is a zinc-dependent peptide chain endonuclease with the ability to degrade the extracellular matrix, and a key factor involved in tumor invasion and metastasis (24-26). MMP-9 is secreted by tumor cells and various immune cells in the stroma. The products formed by MMP-9 activation can decompose the majority of substances present in the basement membrane and extracellular matrix (27). In addition, angiogenesis is increased by the presence of VEGF in tumors (28). Together, the combined function of MMP-9 and VEGF promote tumor invasion and metastasis (29). In the coculture, HPCs significantly upregulated the expression of MMP-9 at the mRNA and protein expression levels in the CCSC subsets. Therefore, it is hypothesized that the local microenvironment formed by the interactions between HPCs and premetastatic tissues results in recruitment of distant CCSC subsets, which promotes the secretion of MMP-9 by tumor cells to accelerate the process of tumor cell metastasis.
Honguero Martínez et al. showed that the invasion, growth and metastasis of primary tumors also requires an abundant blood supply (30). Based on the current body of knowledge, VEGF is the most important angiogenic stimulating factor (31). VEGF primarily promotes tumor metastasis in two ways: it specifically binds and stimulates the proliferation of vascular endothelial cells to induce angiogenesis; and enhances a series of specific signaling pathways which increase the permeability of microvessels, making it easier for tumor cells to penetrate the vascular wall and enter the circulatory system (32). Zheng et al. confirmed that VEGF mRNA expression is upregulated in various types of malignant tumors, and its expression is positively correlated with tumor metastasis (33). In the present study, the mRNA and protein expression levels of VEGF in CCSC subgroup cells were significantly upregulated in the cocultured cells. Thus, it is hypothesized that when tumor cells in the circulatory system reach the target organ, the HPCs send specific stimulation signals to promote CCSC subsets to secrete VEGF, promoting the occurrence of neoplastic angiogenesis, and thus facilitating secondary metastasis.
It is well known that epithelial-mesenchymal transition (EMT) play an important role in colon cancer metastatic progression (34). Firstly, Epithelial cells lose the junctions and apical-basal polarity, then re-arrange the cytoskeletons. This procession up-regulates the motility of individual cells and promotes them to leave the tissue parenchyma, which facilitates the cancer metastasis (34). The hallmarks of EMT include the reduced expression of E-cadherin, cytokeratins and tight junction proteins, in which E-cadherin has a critical role in the whole procession (35). Furthermore, a variety of signaling pathways are associated with the activation of EMT, including Wnt/β-catenin, tumor growth factor-β, RTK/Ras pathways and others (36). The Wnt/β-catenin signaling pathway has been revealed to have important roles in the promotion of EMT in colon cancer metastasis. β-catenin promotes the EMT by binding to the TCF/LEF transcription factors to trans-activate target genes. In our study, we found that HPCs significantly increase the β-catenin expression levels in CCSC subsets. Moreover, E-cadherin expression was remarkably decreased in CCSC subsets. Thus, our results showed that HPCs might promote CCSC metastasis partly by facilitating the EMT procession though increasing β-catenin expression.
Conclusions
In conclusion, through the HPC and CCSC subsets that were studied in an in vitro coculture, we showed that HPCs enhanced the biological properties of CCSCs, including their migratory, invasive and proliferative capacity. The underlying mechanism may be the activation of specific signaling pathways that increases the secretion of specific chemokines, cytokines and protein enzymes. In the present study, HCT116 colon cancer cells were used to study the effect of HPCs on metastasis of colon cancer cells. Similarly, to examine the effect of HPCs on breast cancer metastasis, Meng et al. determined the metastatic ability of MDA-MB-435s-hm cells co-cultured with HPC (37). Although the specific pathways and mechanism of action are not fully understood, it was shown that tumor metastasis was related both to the characteristics of tumor cells, and also the premetastatic microenvironment in which HPCs are involved in generating. Therefore, further research on the mechanisms related to the establishment of a suitable premetastatic microenvironment for colorectal cancer cells may result in the development of novel methods for the treatment of liver metastasis of colorectal cancer, and the development of a series of targeted drugs, in turn, improving the outcomes of CRLM patients.
Acknowledgments
Funding: This study was sponsored by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-2810
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2810
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2810). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- Ochiai T, Masuda T, Yagi M, et al. Successful combination therapy of radical liver resection with 5-fluorouracil/leucovorin, oxaliplatin, plus bevacizumab for ascending colon cancer with pulmonary and 43 liver metastases: report of a case. Int Surg 2012;97:6-13. [Crossref] [PubMed]
- Meza R, Jeon J, Renehan AG, et al. Colorectal cancer incidence trends in the United States and United kingdom: evidence of right- to left-sided biological gradients with implications for screening. Cancer Res 2010;70:5419-29. [Crossref] [PubMed]
- Yin Z, Liu C, Chen Y, et al. Timing of hepatectomy in resectable synchronous colorectal liver metastases (SCRLM): Simultaneous or delayed? Hepatology 2013;57:2346-57. [Crossref] [PubMed]
- Paget S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev 1989;8:98-101. [PubMed]
- Cabrera MC, Hollingsworth RE, Hurt EM. Cancer stem cell plasticity and tumor hierarchy. World J Stem Cells 2015;7:27-36. [Crossref] [PubMed]
- Soltanian S, Matin MM. Cancer stem cells and cancer therapy. Tumour Biol 2011;32:425-40. [Crossref] [PubMed]
- Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov 2014;13:497-512. [Crossref] [PubMed]
- Sleeman JP. The metastatic niche and stromal progression. Cancer Metastasis Rev 2012;31:429-40. [Crossref] [PubMed]
- Chin AR, Wang SE. Cancer tills the premetastatic field: mechanistic basis and clinical implications. Clin Cancer Res 2016;22:3725-33. [Crossref] [PubMed]
- Kaplan RN, Riba RD, Zacharoulis S, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 2005;438:820-827. [Crossref] [PubMed]
- Zhang C, Zhou C, Wu XJ, et al. Human CD133-positive hematopoietic progenitor cells initiate growth and metastasis of colorectal cancer cells. Carcinogenesis 2014;35:2771-7. [Crossref] [PubMed]
- García-Gómez I, Elvira G, Zapata AG, et al. Mesenchymal stem cells: biological properties and clinical applications. Expert Opin Biol Ther 2010;10:1453-68. [Crossref] [PubMed]
- Song DY, Wang Y, Sun L, et al. Progress in tumor-stem cell theory and tumor stem cells isolation and identification. Chinese Journal of Pharmacology and Toxicology 2012;26:674-9.
- Martens DJ, Tropepe V, van Der Kooy D. Separate proliferation kinetics of fibroblast growth factor-responsive and epidermal growth factor-responsive neural stem cells within the embryonic forebrain germinal zone. J Neurosci 2000;20:1085-95. [Crossref] [PubMed]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature 2007;445:111-5. [Crossref] [PubMed]
- Vermeulen L, Todaro M, de Sousa Mello F, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 2008;105:13427-32. [Crossref] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Luzzi KJ, MacDonald IC, Schmidt EE, et al. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol 1998;153:865-73. [Crossref] [PubMed]
- Psaila B, Kaplan RN, Port ER, et al. Priming the ‘soil’ for breast cancer metastasis: the pre-metastatic niche. Breast Dis 2006-2007;26:65-74. [Crossref] [PubMed]
- Jain S, Ward MM, O’Loughlin J, et al. Incremental increase in VEGFR1 + hematopoietic progenitor cells and VEGFR2 + endothelial progenitor cells predicts relapse and lack of tumor response in breast cancer patients. Breast Cancer Res Treat 2012;132:235-42. [Crossref] [PubMed]
- Hur K, Toiyama Y, Okugawa Y, et al. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut 2017;66:654-65. [Crossref] [PubMed]
- Crotti S, Piccoli M, Rizzolio F, et al. Nitti D and Agostini M. Extracellular matrix and colorectal cancer: how sur-rounding microenvironment affects cancer cell behavior? J Cell Physiol 2017;232:967-75. [Crossref] [PubMed]
- Zheng CG, Chen R, Xie JB, et al. Immunohistochemical expression of Notch1, Agged1, NF-κB and MMP-9 in colorectal cancer patients and the relationship to clinicopathological parameters. Cancer Biomark 2015;15:889-97. [Crossref] [PubMed]
- Han J, Tian R, Yong B, et al. Gas6/Axl mediates tumor cell apoptosis, migration and invasion and predicts the clinical outcome of osteosarcoma patients. Biochem Biophys Res Commun 2013;435:493-500. [Crossref] [PubMed]
- Czyzewska J, Guzińskaustymowicz K, Kemona A, et al. The expression of matrix metalloproteinase 9 and cathepsin B in gastric carcinoma is associated with lymph node metastasis, but not with postoperative survival. Folia Histochem Cytobiol 2008;46:57-64. [Crossref] [PubMed]
- Zhang S, Mesalam A, Joo MD, et al. Matrix metalloproteinases improves trophoblast invasion and pregnancy potential in mice. Theriogenology 2020;151:144-50. [Crossref] [PubMed]
- Viloria-Petit A, Miquerol L, Yu JL, et al. Contrasting effects of VEGF gene disruption in embryonic stem cell-derived versus oncogene-induced tumors. EMBO J 2003;22:4091-102. [Crossref] [PubMed]
- Celus W, Di Conza G, Oliveira AI, et al. Loss of Caveolin-1 in Metastasis-Associated Macrophages Drives Lung Metastatic Growth through Increased Angiogenesis. Cell Rep 2017;21:2842-54. [Crossref] [PubMed]
- Honguero Martínez AF, Arnau Obrer A, Figueroa Almazán S, et al. Prognostic value of the expression of vascular endothelial growth factor A and hypoxia-inducible factor 1alpha in patients undergoing surgery for non-small cell lung cancer. Med Clin (Barc) 2014;142:432-7. [PubMed]
- Frezzetti D, Gallo M, Roma C, et al. Vascular Endothelial Growth Factor A Regulates the Secretion of Different Angiogenic Factors in Lung Cancer Cells. J Cell Physiol 2016;231:1514-21. [Crossref] [PubMed]
- Zhu YS, Li YM, Lu HS, et al. Relationship between vascular endothelial growth factor and invasion and metastasis of colorectal cancer. Chinese Journal of Celiopathy 2004;4:84-6.
- Zheng Y, Hao Z, Ding Y, et al. Expression of delta-like 4 (Drosophila) and vascular endothelial growth factor A in colon cancer and association with tumour angiogenesis. J Int Med Res 2015;43:535-43. [Crossref] [PubMed]
- Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol 2007;19:150-8. [Crossref] [PubMed]
- Lai-Cheong JE, Parsons M, Tanaka A, et al. Loss-of-function FERMT1 mutations in kindler syndrome implicate a role for fermitin family homolog-1 in integrin activation. Am J Pathol 2009;175:1431-41. [Crossref] [PubMed]
- Li VS, Ng SS, Boersema PJ, et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 2012;149:1245-56. [Crossref] [PubMed]
- Meng D, Meng M, Luo A, et al. Effects of VEGFR1+ hematopoietic progenitor cells on pre-metastatic niche formation and in vivo metastasis of breast cancer cells. J Cancer Res Clin Oncol 2019;145:411-27. [Crossref] [PubMed]


