
SAV1, regulated by HERC4, inhibits the proliferation, migration, and invasion of hepatocellular carcinoma
Introduction
Primary hepatic carcinoma is one of the most malignant s cancers worldwide (1). Hepatocellular carcinoma (HCC) accounts for 70% to 90% of primary hepatic carcinoma (2). It has the fifth highest incidence of cancer in the world and the third highest death rate (3). There were 782,000 new cases worldwide and 745,000 patients died in 2012 (4). Currently, surgical resection is the preferred treatment for HCC, while chemoradiotherapy, interventional therapy, and immunotherapy are not preferred treatments (5,6). However, the clinical diagnosis is still dominated by advanced HCC due to the insidious onset of HCC and the absence of obvious early symptoms. Only 10–15% of patients with advanced HCC are suitable for surgical treatment, and the 5-year survival rate is only 5–9%, while the 5-year survival rate of patients who are diagnosed early and undergo hepatectomy increase to 69% (7,8). How to effectively prevent or delay the progression of HCC is a hot and difficult topic in current studies. Therefore, it is of great significance to further explore the pathogenesis of HCC and identify the key targets for regulating the HCC process.
Abnormal gene expression and dysregulation of signal transduction networks can generate pathological proliferation, apoptosis, migration, and invasion of cells, and further accelerate cancer development (9-11). The ubiquitin proteasome system (UPS) has been found to be essential in regulating the dynamic balance of proteins in vivo and thereby affecting a series of physiological processes (12,13). In addition, growing evidence has suggested that UPS dysfunction is closely related to the development of cancer (14,15). The E3 ubiquitin ligase is an enzyme with specific recognition of substrate proteins, and is significant part of the ubiquitination pathway (16,17). The E3 ubiquitin ligase is mainly divided into three types: HECT, TING, and u-box domain families (18). Among them, HERC4 belongs to the HECT family, and its gene is located on chromosome 10q21.3. Findings have suggested that HERC4 has significant clinical significance in several types of cancers including cervical cancer (19), lung cancer (20), and breast cancer (21,22). Moreover, research has also confirmed that HERC4 can expedite proliferation and migration of HCC (23). However, the downstream regulatory mechanism of HERC4 in HCC has not been elucidated.
The Hippo signaling pathway has been actively studied in recent years as it affects cell proliferation, apoptosis, and metastasis (24). Also, it has an important regulatory function in the development of cancer, stem cell function, tissue regeneration, and cell contact inhibition (25-28). Salvador 1 (SAV1, also known as WW45), a core kinase component of the Hippo signaling pathway, plays an extensive and prominent role in a variety of human carcinogenesis (29,30). For instance, SAV1 can inhibit tumor growth of colorectal cancer (29) and a decrease of SAV1 can accelerate the pathological progression of high-grade clear cell renal cell carcinoma (30). However, the possible mechanism and related function of SAV1 in HCC is not yet clearly established. Our preliminary experiment found that HERC4 could down-regulate the level of SAV1 in HCC. However, it is not clear whether HERC4 can directly bind to SAV1 and participate in HCC processes.
We further identified the levels and regulatory relationships of HERC4 and SAV1 in HCC. In addition, we investigated the influences of SAV1 overexpression on proliferation, migration, and invasion of HCC by being regulated by HERC4. Therefore, it is an extremely attractive hypothesis that HERC4 and SAV1 might be new therapeutic targets for HCC.
Methods
HCC tumor samples
HCC and para-carcinoma non-tumor tissues (15 pairs) were harvested from HCC patients who were diagnosed at the Second Affiliated Hospital of Zhejiang Chinese Medical University from May 2017 to April 2018. These samples were immediately saved in liquid nitrogen until use. We also obtained informed consents, which were provided by each participant, and our study was approved by the ethics committee of The Second Affiliated Hospital of Zhejiang Chinese Medical University (2018-KL-020-01). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture
Hep 3B cells (BNCC352197) were obtained from the BeNa Culture Collection (Beijing, China) and grown in MEM (Gibco) containing 10% fetal bovine serum (FBS; Gibco, cat. no. 26140079) with 1% Penicillin-Streptomycin Solution at 37 °C and 5% CO2.
Plasmid construction and transfection
SAV1 and HERC4 overexpression plasmids were purchased from Genomeditech Company (Shanghai, China). HEP 3B cells (1×105 cells/well) were plated in 6-well plates and incubated for 8 h at 37 °C. Next, these cells were transfected with empty vector, SAV1-overexpressing plasmids and HERC4-overexpressing plasmids.
RNA extraction and quantitative real-time PCR (RT-qPCR) assay.
HCC and para-carcinoma tissues were ground at a low temperature. TRIzol reagent (Invitrogen, USA) was used to extract total RNAs from the ground tissues and the treated HEP 3B cells according to the instruction. The obtained RNAs were quantitatively determined using a NanoDro2000c (Thermo Scientific). 1.0 µg RNA was used to synthesize cDNA using the reverse transcription kit (Takara, Japan). The expression level of each indicator was examined and quantified using BestarTM qPCR MasterMix (DBI Bioscience) based on the specification provided by the supplier. GAPDH was used as an internal control. The sequences of primers are listed in Table 1.
Table 1
| Gene | Sequence or target sequence |
|---|---|
| GAPDH-F | CACCCACTCCTCCACCTTTG |
| GAPDH-R | CCACCACCCTGTTGCTGTAG |
| HERC4-F | TGGAATCCCTTTCATGCAAGTT |
| HERC4-R | TCCTTGGTTAGAGCAGCAGTAT |
| SAV1-F | ATGAGGCGTGAAAGCAACAG |
| SAV1-R | CCGCTGTGCTCATAGTATCTGTA |
F, forward primer; R, reversed primer.
Western blotting analysis
The ground tissues and the treated HEP 3B and Huh7 cells were combined with moderate ice-cold RIPA buffer (Santa Cruz) supplemented g with the protease inhibitor cocktail (Sigma). After centrifugation (12,000 ×g for 30 min at 4 °C), the concentration of protein was quantitively determined using the bicinchoninic acid (BCA) kit (Beyotime Biotechnology). A total of 30 µg protein was isolated by 10% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad). After sealing with 5% skim milk, the membranes were treated with primary antibodies against anti-HERC4 (1:1,000; Abcam, ab221757), anti-SAV1 (1:1,000; Abcam, ab105105), and caspase 3 (1:500. Abcam, ab32042). Then horseradish peroxidase-conjugated secondary antibodies were used to treat the blots for 1 h. The blots were visualized using enhanced chemiluminescence (ECL; Bio-Rad, USA).
Co-immunoprecipitation (Co-IP) assay
HEP 3B and Huh7 cells were added with the cell lysis solution containing the protease inhibitor and incubated for 30 min on ice. After centrifugation (12,000 ×g for 30 min at 4 °C), an adequate amount of the protein was used as the input group, the remaining proteins were incubated with 2 µg of the corresponding antibodies (anti-HERC4) and anti-IgG (negative control) for 1 h at 4 °C. and the proteins were added to protein A-Agarose at 4 °C incubation overnight. After washing with lysates, the beads were centrifuged (2,500 ×g for 2 min at 4 °C). After boiling denaturation with the loading buffer, the proteins were used for Western blot analysis.
Immunofluorescence (IF) assay
The treated HEP 3B and Huh7 cells were spread on slides and incubated for 8 h at 37 °C. The adhering cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 30 mins. After sealing with 10% normal goat serum for 30 mins, cells were incubated with the primary antibodies (anti-SAV1; 1 µg/mL, Abcam, ab105105) at 4 °C overnights. The cells were treated with fluorescent labeled goat anti-rabbit IgG (1:200) at room temperature for 1 h in the dark. After treatment with an anti-fluorescent quenching agent, they were observed and photographed under a fluorescent microscope.
Edu staining
The treated HEP 3B and Huh7 cells (2×104 cells/well) were put into 24-well plates and 300 µL Edu solution was added (50 µmol/L) for 1 h. After washing, cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 30 min and 2 mg/mL glycine was added for 5 min. After washing, cells were treated with 300 µL permeating agent including 0.5% Triton X-100 for 10 min and 300µL DAPI for 10 mins. The staining results were observed under a microscope.
Colony formation assay
HEP 3B and Huh7 cells were placed in 35-mm culture dishes at a concentration of 3000 cells/dish. After transfection with SAV1 or/and HERC4-overexpressing plasmids, cells were grown for two weeks at 37 °C with 5% CO2. The colonies were fixed in 4% paraformaldehyde for 10 min and stained with Giemsa solution for 10 mins. The number of colonies were counted under a microscope.
Transwell assay
For the migration assay, the transfected HEP 3B cells were counted, and a single cell suspension (5×105 cells/ml) was prepared using serum free medium after pancreatic enzyme digestion. 200 µL cell suspensions were inoculated into the upper Transwell chamber, and 600 µL medium containing 15%FBS was placed into the lower Transwell chamber. After incubation for 24 h at 37 °C, the migrated cells were fixed using 4% paraformaldehyde (Sigma-Aldrich) for 10 min and dyed with 0.1% crystal violet for 20 min. After washing, the migrated cells were observed using a microscope. For the invasion assay, Matrigel was applied on the Transwell chamber and incubated for 30 min at 37 °C, while the other processes were the same as the migration assay.
In vivo xenograft tumor-burdened model
A total of 10 female Balb/c nude mice (weight: 14–17 g) were purchased from Zhejiang Chinese Medical University Laboratory Animal Research Center and then maintained in specific pathogen-free atmosphere. All mice have access to sterilized food and water. Mice were then injected subcutaneously with HEP3B cells (106 cells each mouse) after one week of adjustable feeding. 30 days later, all the mice were sacrificed and tumor tissues were collected for size measurement and low temperature preservation. Tumor volume was calculated using formula: tumor volume (mm3)= [tumor length × (tumor width)2]/2. All animals experiments were conducted according to institutional guideline and approved by Ethics Committee of The Second Affiliated Hospital of Zhejiang Chinese Medical University.
Immunohistochemistry
Tumor tissues were maintained in 4% paraformaldehyde for fixation followed by paraffin-embedding. Paraffin-embedded tissues serial sections (4 µm) were obtained, and then dewaxing and dehydration with xylene and hydrated respectively. Sections were incubated with primary antibody (1:200, PB9026, Boster, China) overnight at 4o °C after antigen retrieval and sealing. Afterwards, sections were incubated with horseradish peroxidase-conjugated secondary antibody (1:200, BM3895, Boster, China) at room temperature. Sections were developed with diaminobenzidine (DAB) and hematoxylin. Finally, sections sealed with neutral resin were observed under microscope.
Statistical Analysis
GraphPad Prism Software (Ver. Prism 7) was used for statistical analysis of data, all of which were expressed as mean ± SD. The Student t-test was used to calculate the difference between the two groups. The overall survival was obtained from Kaplan-Meier plots. P<0.05 was considered statistically significant.
Results
HERC4 was highly expressed, while SAV1 was lowly expressed in HCC tissues
To verify expression changes of HERC4 and SAV1, we extracted the total mRNA and protein from 15 pairs of HCC and para-carcinoma tissues. As shown in Table 2, the clinical feature of tumor size and TNM stage of patients presented significant difference with expression of SAV1. However, HERC4 expression displayed no significant difference with tumor size (P=0.13). This discrepancy might due to a small number of involved subjects (Table 2).
Table 2
| Features | N | SAV1 expression | HERC4 expression | |||||
|---|---|---|---|---|---|---|---|---|
| High | Low | P value | High | Low | P value | |||
| Age | ||||||||
| <53 | 8 | 2 | 6 | 0.31 | 5 | 3 | 0.31 | |
| ≥53 | 7 | 5 | 2 | 2 | 5 | |||
| Gender | ||||||||
| Male | 9 | 4 | 5 | 0.90 | 3 | 6 | 0.31 | |
| Female | 6 | 3 | 3 | 4 | 2 | |||
| Tumor size | ||||||||
| <5 cm | 8 | 6 | 2 | 0.04 | 2 | 6 | 0.13 | |
| ≥5 cm | 7 | 1 | 6 | 5 | 2 | |||
| TNM stage | ||||||||
| I + II | 7 | 6 | 1 | 0.01 | 1 | 6 | 0.04 | |
| III + IV | 8 | 4 | 7 | 6 | 2 | |||
SAV1, Salvador 1.
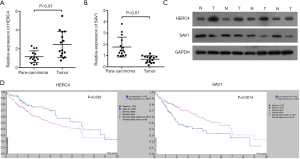
The results from the RT-qPCR assay showed that the level of HERC4 was markedly elevated in HCC tissue samples relative to para-carcinoma tissues (P<0.01, Figure 1A). Our results showed that SAV1 expression was significantly lower in HCC samples than that in para-carcinoma ones (P<0.01, Figure 1B). In addition, Western blot results also confirmed high expression of HERC4 and low expression of SAV1 in HCC tissues (Figure 1C). More importantly, data from Kaplan-Meier plots disclosed that HCC patients with high HERC4 expression exhibited a worse prognosis than those HCC patients with low HERC4, while HCC patients with high SAV1 expression had a longer survival trend than those HCC patients with low SAV1 expression (Figure 1D). Therefore, these data implied that HERC4 and SAV1 might make significant contributions during HCC tumorigenesis.
SAV1 significantly prevented HCC cell proliferation through interaction with HERC4
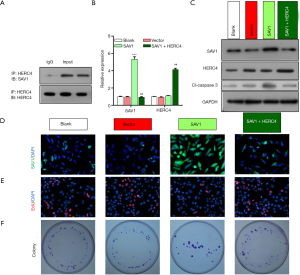
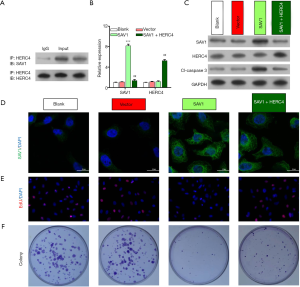
Because of the opposite regulatory trends of HERC4 and SAV1 in HCC tissues, we further verified whether HERC4 was a direct target of SAV1 in HEP3B and Huh7 cells. Firstly, the Co-IP assay with anti-HERC4 antibodies was used to verify the interaction between HERC4 and SAV1. As shown in Figure 2 and Figure 3, the results from the CO-IP assay proved the formation of a complex between HERC4 and SAV1 in HEP 3B cells (Figure 2A) and Huh7 cells (Figure 3A), suggesting that HERC4 can directly interact with SAV1 in HCC cells. In order to further verify the regulatory effect of SAV1 on HERC4 in HCC cells, HEP 3B cells were overexpressed with SAV1, and SAV1-overexpressing HEP 3B cells were then transfected with HERC4 overexpressing plasmids. Results showed that the level of SAV1 was prominently upregulated in HEP 3B and Huh7 cells after overexpression of SAV1, while this upregulation was completely reversed by HERC4 overexpression. Also, it was found that overexpression of HERC4 could markedly improve the expression level HERC4 in HCC cells (P<0.01, P<0.001, Figure 2B,C, Figure 3B,C). In addition, the results of IF assay also demonstrated the inhibitory action of HERC4 on SAV1 (Figure 2D, Figure 3D). We further investigated the possible effects of HERC4 and SAV1 on HCC cell proliferation, and the results from Edu staining and colony formation assay showed that overexpression of SAV1 could inhibit HEP 3B and Huh7 cell proliferation, while this inhibiting effect could be almost completely reversed by HERC4 overexpression, indicating that HERC4-inhibition of SAV1 was involved in the proliferation of HCC cells (Figure 2E,F, Figure 3E,F). In consequence, we proposed that the interaction of HERC4 and SAV1might contribute to the tumorigenesis of HCC.
Overexpression of SAV1 suppressed the migration and invasion of HCC cells by being regulated by HERC4
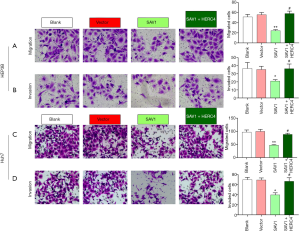
To further verify whether HERC4 could regulate the role of SAV1 on the migration and invasion of HCC cells, we co-transfected the overexpressing plasmids of HERC4 and SAV1 into HEP 3B cells. Transwell results showed that the migration and invasion of HEP 3B cells could be significantly suppressed by overexpression of SAV1, while overexpression of HERC4 then dramatically reversed this suppression mediated by SAV1 (P<0.05, P<0.01, Figure 4). As a consequence, it was found that HERC4 could participate in the migration (Figure 4A,C) and invasion (Figure 4B,4D) of HCC cells by downregulating SAV1 in HEP3B and Huh7 cells.
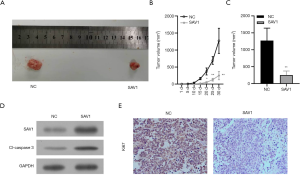
Overexpression of SAV1 inhibited tumor growth in nude mice
To confirm the tumor-inhibitive effect of SAV1 in HCC, experiment in vivo involving nude mice with a tumor-burdened was performed. As shown in Figure 5, SAV1 overexpression significantly suppressed tumor growth (Figure 5A,B,C). Further investigation showed that SAV1 overexpression enhanced caspase 3 expression in tumor compared with NC (Figure 5D), together with weaker expression of Ki67 in tumor, as shown in Figure 5E.
Discussion
The Hippo pathway, a cascade enzyme chain reaction composed of a series of protein kinases, is a newly elaborated cell signaling pathway (31). The Hippo pathway is highly genetically conserved, and mainly contains Mst1/2, SAV1, lats1/2, and Mob1 proteins (32). The Hippo pathway is considered as a tumor suppressor pathway, and the multiple core proteins of it are tumor suppressors (33). Studies have discovered that the inactivation of the Hippo pathway is connected with the occurrence and development of cancers (34,35). Among them, the WW and SARAH domains of SAV1 can interact with other proteins (36). In the Hippo pathway, SAV1 can act as a bridging protein between Mst1/2 and Lats1/2 (37). In addition, SAV1 can enhance Mst1 and Mst2 to induce apoptosis (38). Current research also suggested that SAV1 can prevent the development of colorectal cancer (39). In vivo experiments proved that the knockout of SAV1 can cause a liver volume increase and the formation of liver cancer. Therefore, SAV1 is thought to be a tumor-suppressor gene that can repress the proliferation of cancer cells. However, existing studies have shown little understanding of SAV1, and its biological function is not fully explained in HCC. In this report, we emphasized that the level of SAV1 was lowly expressed in HCC, and patients with high SAV1 had a good prognosis. It was found that overexpression of SAV1 significantly prevented the proliferation and colony formation capacity of HEP 3B cells. Also, we found that overexpression of SAV1 could dramatically result in the suppressions of migration and invasion of HEP 3B cells. Therefore, our data provide a strong hypothesis that SAV1 may be a potential suppressor gene for HCC.
The initiation of cancer is related to gene mutation, gene expression disorder, and protein regulation disorder (40). In HCC, mutations or expression disorders of key regulatory genes may result in the occurrence, development, and metastasis of cancers (41). Studies have proved that UPS is closely associated with the occurrence of cancer (42,43). Furthermore, its members can not only serve as potential cancer diagnostic markers or prognostic indicators, but also as potential molecular therapeutic targets. At present, E3 ubiquitin ligase in UPS is widely studied because of its specificity (43). Of note, HERC4, as a newly identified E3 ubiquitin ligase, has been shown to contribute to cancer progression and affect prognosis (44). For example, upregulation of HERC4 is positively related to the histological grade, TNM stage, and metastasis of lung cancer (20). HERC4 is associated with the progression of breast cancer (21), and contributes to the tumorigenesis of breast cancer (45). HERC4 has also been proved to participate in the proliferation, apoptosis, and migration of cervical cancer (19). HERC4 can prevent the growth of myeloma xenografts by regulating c-Maf (46). In our study, the upregulation of HERC4 was validated in HCC tissue samples, and patients with high HERC4 exhibited a worse prognosis. In addition, our study is the first report demonstrating the targeted regulation between HERC4 and SAV1, SAV1 was identified as a target gene of HERC4, and it can be negatively regulated by HERC4 in HCC. Moreover, functional experiments have confirmed that HERC4 can reverse the inhibiting effects of SAV1 on HCC cell proliferation, migration, and invasion. Therefore, the oncogenic role of HERC4 on HCC progress could be achieved by suppressing expression of SAV1.
In conclusion, by employing an overexpression of SAV1 in the HCC cell line, cell proliferation, migration, and invasion was suppressed. However, HERC4 overexpression rescued the inhibitory effect of SAV1 and cell proliferation, migration, and invasion were the enhanced. This study provided evidences that SAV1, an antioncogene of HCC, can be repressed by HERC4. This study suggested that HERC4 can be a potential target of HCC.
Acknowledgments
We thank to Dr. Edward C. Mignot, Shandong University, for linguistic advice.
Funding: This research was financially supported by
Footnote
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-698
Conflicts of Interests: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-698). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by The Second Affiliated Hospital of Zhejiang Chinese Medical University (2018-KL-020-01), and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sui W, Gan Q, Liu F, Chen H, Liu J, Dai Y. The differentially expressed circular ribonucleic acids of primary hepatic carcinoma following liver transplantation as new diagnostic biomarkers for primary hepatic carcinoma. Tumour Biol 2018;40:1010428318766928. [Crossref] [PubMed]
- Budny A, Kozlowski P, Kaminska M, et al. Epidemiology and risk factors of hepatocellular carcinoma. Pol Merkur Lekarski 2017;43:133-9. [PubMed]
- Xiong XX, Qiu XY, Hu DX, et al. Advances in Hypoxia-Mediated Mechanisms in Hepatocellular Carcinoma. Mol Pharmacol 2017;92:246-55. [Crossref] [PubMed]
- Xia J, Song P, Sun Z, et al. Advances of diagnostic and mechanistic studies of gamma-glutamyl transpeptidase in hepatocellular carcinoma. Drug Discov Ther 2016;10:181-7. [Crossref] [PubMed]
- Akoad ME, Pomfret EA. Surgical resection and liver transplantation for hepatocellular carcinoma. Clin Liver Dis 2015;19:381-99. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Kokudo T, Hasegawa K, Matsuyama Y, et al. Survival benefit of liver resection for hepatocellular carcinoma associated with portal vein invasion. J Hepatol 2016;65:938-43. [Crossref] [PubMed]
- Agrawal S, Agarwal S, Arnason T, et al. Management of Hepatocellular Adenoma: Recent Advances. Clin Gastroenterol Hepatol 2015;13:1221-30. [Crossref] [PubMed]
- Scarabel L, Toffoli G. Does PD1/PDL1 Immunotherapy Require Patient Tumor Subgroup Pre-Analysis in the Era of Precision Medicine? J Mol Med Clin Med 2018;1:51-4.
- Bradner JE, Hnisz D, Young RA. Transcriptional Addiction in Cancer. Cell 2017;168:629-43. [Crossref] [PubMed]
- Mahale A, Alkatan H, Alwadani S, et al. Altered gene expression in conjunctival squamous cell carcinoma. Mod Pathol 2016;29:452-60. [Crossref] [PubMed]
- Kwon YT, Ciechanover A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci 2017;42:873-86. [Crossref] [PubMed]
- Nandi D, Tahiliani P, Kumar A, et al. The ubiquitin-proteasome system. J Biosci 2006;31:137-55. [Crossref] [PubMed]
- Ao N, Chen Q, Liu G. The Small Molecules Targeting Ubiquitin-Proteasome System for Cancer Therapy. Comb Chem High Throughput Screen 2017;20:403-13. [Crossref] [PubMed]
- Shen M, Schmitt S, Buac D, et al. Targeting the ubiquitin-proteasome system for cancer therapy. Expert Opin Ther Targets 2013;17:1091-108. [Crossref] [PubMed]
- Peng J, Ren KD, Yang J, et al. Mitochondrial E3 ubiquitin ligase 1: A key enzyme in regulation of mitochondrial dynamics and functions. Mitochondrion 2016;28:49-53. [Crossref] [PubMed]
- Shearer RF, Iconomou M, Watts CK, et al. Functional Roles of the E3 Ubiquitin Ligase UBR5 in Cancer. Mol Cancer Res 2015;13:1523-32. [Crossref] [PubMed]
- McDermott JE, Cort JR, Nakayasu ES, et al. Prediction of bacterial E3 ubiquitin ligase effectors using reduced amino acid peptide fingerprinting. PeerJ 2019;7:e7055. [Crossref] [PubMed]
- Wei M, Zhang YL, Chen L, et al. Nan Fang Yi Ke Da Xue Xue Bao 2016;37:232-7. [RNA interference of HERC4 inhibits proliferation, apoptosis and migration of cervical cancer Hela cells]. [PubMed]
- Zeng WL, Chen YW, Zhou H, et al. Expression of HERC4 in lung cancer and its correlation with clinicopathological parameters. Asian Pac J Cancer Prev 2015;16:513-7. [Crossref] [PubMed]
- Zhou H, Shi R, Chen Y, et al. Nan Fang Yi Ke Da Xue Xue Bao 2014;34:1110-4. [Expression of E3 ligase HERC4 in breast cancer and its clinical implications]. [PubMed]
- Zhou H, Shi R, Wei M, et al. The expression and clinical significance of HERC4 in breast cancer. Cancer Cell Int 2013;13:113. [Crossref] [PubMed]
- Zheng Y, Li J, Pan C, et al. HERC4 Is Overexpressed in Hepatocellular Carcinoma and Contributes to the Proliferation and Migration of Hepatocellular Carcinoma Cells. DNA Cell Biol 2017;36:490-500. [Crossref] [PubMed]
- Janse van Rensburg HJ, Yang X. The roles of the Hippo pathway in cancer metastasis. Cell Signal 2016;28:1761-72. [Crossref] [PubMed]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010;19:491-505. [Crossref] [PubMed]
- Wierzbicki PM, Rybarczyk A. The Hippo pathway in colorectal cancer. Folia Histochem Cytobiol 2015;53:105-19. [Crossref] [PubMed]
- Hong AW, Meng Z, Guan KL. The Hippo pathway in intestinal regeneration and disease. Nat Rev Gastroenterol Hepatol 2016;13:324-37. [Crossref] [PubMed]
- Wang Y, Yu A, Yu FX. The Hippo pathway in tissue homeostasis and regeneration. Protein Cell 2017;8:349-59. [Crossref] [PubMed]
- Jiang J, Chang W, Fu Y, et al. SAV1, regulated by microRNA-21, suppresses tumor growth in colorectal cancer. Biochem Cell Biol 2019;97:91-9. [Crossref] [PubMed]
- Matsuura K, Nakada C, Mashio M, et al. Downregulation of SAV1 plays a role in pathogenesis of high-grade clear cell renal cell carcinoma. BMC Cancer 2011;11:523. [Crossref] [PubMed]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer 2013;13:246-57. [Crossref] [PubMed]
- Ercolani C, Benedetto AD, Terrenato I, et al. Expression of phosphorylated Hippo pathway kinases (MST1/2 and LATS1/2) in HER2-positive and triple-negative breast cancer patients treated with neoadjuvant therapy. Cancer Biol Ther 2017;18:339-46. [Crossref] [PubMed]
- Furth N, Aylon Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ 2017;24:1488-501. [Crossref] [PubMed]
- Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015;163:811-28. [Crossref] [PubMed]
- Gregorieff A, Wrana JL. Hippo signalling in intestinal regeneration and cancer. Curr Opin Cell Biol 2017;48:17-25. [Crossref] [PubMed]
- Kai T, Tsukamoto Y, Hijiya N, et al. Kidney-specific knockout of Sav1 in the mouse promotes hyperproliferation of renal tubular epithelium through suppression of the Hippo pathway. J Pathol 2016;239:97-108. [Crossref] [PubMed]
- Meng Z, Moroishi T, Mottier-Pavie V, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 2015;6:8357. [Crossref] [PubMed]
- Chang Y, Fu XR, Cui M, et al. Activated hippo signal pathway inhibits cell proliferation and promotes apoptosis in NK/T cell lymphoma cells. Cancer Med 2019;8:3892-904. [Crossref] [PubMed]
- Jiang J, Chang W, Fu Y, et al. SAV1 represses the development of human colorectal cancer by regulating the Akt-mTOR pathway in a YAP-dependent manner. Cell Prolif 2017;50:e12351. [Crossref] [PubMed]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 2008;9:759-69. [Crossref] [PubMed]
- Calderaro J, Couchy G, Imbeaud S, et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol 2017;67:727-38. [Crossref] [PubMed]
- Lub S, Maes K, Menu E, et al. Novel strategies to target the ubiquitin proteasome system in multiple myeloma. Oncotarget 2016;7:6521-37. [Crossref] [PubMed]
- Johnson DE. The ubiquitin-proteasome system: opportunities for therapeutic intervention in solid tumors. Endocr Relat Cancer 2015;22:T1-17. [Crossref] [PubMed]
- Mao X, Sethi G, Zhang Z, et al. The Emerging Roles of the HERC Ubiquitin Ligases in Cancer. Curr Pharm Des 2018;24:1676-81. [Crossref] [PubMed]
- Xu Y, Ji K, Wu M, et al. A miRNA-HERC4 pathway promotes breast tumorigenesis by inactivating tumor suppressor LATS1. Protein Cell 2019;10:595-605. [Crossref] [PubMed]
- Zhang Z, Tong J, Tang X, et al. The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood 2016;127:1676-86. [Crossref] [PubMed]


