
Effects of HOX transcript antisense intergenic RNA on the metastasis, epithelial-mesenchymal transition, and Notch signaling pathway in tongue cancer
Introduction
Squamous cell carcinoma of the tongue (TSCC) is the most common malignancy of the head and neck, and has shown a tendency of increase in prevalence in the young population. To date, the prognosis of tongue cancer is usually affected by lymph node metastasis, which hampers the overall health of patients (1,2). LncRNAs have rised extensive attention as an significant component of gene regulation network, long non-coding RNA HOTAIR (lncHOTAIR) is the first lncRNA recognized as having reversely transcriptional effects (3). Recently, it has been reported to contribute to the pathogenesis of malignancy by inhibiting the anti-oncogene in patients with breast, bladder, and esophageal cancer (4-6). Epithelial-mesenchymal transition (EMT) is the major route of migration and metastasis of cancer cells, including TSCC. According to the previous description, HOTAIR was highly expressed in TSCC tissues compared with the adjacent normal tissues (7). Besides, malignant patients with over-expression of HOTAIR usually show poor prognosis and lymph node metastasis (8). These features led us to investigate the roles of HOTAIR in the pathogenesis of TSCC.
In this study, we explored the lncHOTAIR expression in Tca8113 and TSCCA cells. Besides, we determined the expression of EMT and Notch signaling ligands including Jagged-1, Notch-1 receptor, and downstream Hes-1 protein, to identify whether there was a potential link between RNA HOTAIR and EMT or the Notch signaling pathway.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tcr-20-3452).
Methods
Cells
We purchased TSCCA and Tca8113 cells from the Chinese Academy of Medical Science. Cells were cultured in high glucose medium (HyClone, South Logan, UT, USA) containing 10% fetal bovine serum (FBS, HyClone, South Logan, UT, USA) at 37 °C in 5% CO2. The medium was replaced every 48 h. The study protocols were approved by the Ethical Committee of The First Affiliated Hospital of Xinjiang Medical University (No. K202012-05).
Plasmid construction and transfection
The cells were divided into two groups, an experimental group (siRNA-1 group, siRNA-2 group, siRNA-3) and a negative control group (NC group). The sequences for siRNA-1, siRNA-2, and siRNA-3 were as follows:
5'-CAUCUUGAGACACAUGGGUTT-3' and 5'-ACCCAUGUGUCUCAAGAUGTT-3' (for siRNA-1); 5'-CACUUAAAUAAGUGAUGCATT-3' and 5'-UGCAUCACUUAUUUAAGUGTT-3' (for siRNA-2); and 5'-CUUAUGGAGUAUAUACUCATT-3' and 5'-UGAGUAUAUACUCCAUAAGTT-3' (for siRNA-3). The sequences for NC were 5'-UUCUCCGAACGUGUCACGUTT-3' and 5’-ACGUGACACGUUCGGAGAATT.
Transfection was performed using lipofectamineTM2000 kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Real-time PCR
Total RNA was extracted from the cell lines with Trizol reagent. The complementary (c)DNA synthesis was carried out using a commercial reverse transcription kit (QIAGEN, Hilden, Germany) according to -the instructions of the reagent. Real-time polymerase chain reaction (qPCR) was carried out in a total volume of 20 µL containing 10 µL SybrGreen qPCR mastermix 10 (QIAGEN, Dusseldorf, Germany), 0.5 µL each specific primer, and 1 µL cDNA template. The PCR conditions consisted of denaturation at 94 °C for 2 minutes, followed by 40 cycles of denaturation at 95 °C for 10 seconds, annealing at 56 °C for 30 seconds, and extension at 72 °C for 60 seconds. Primer sequences were as follows: HOTAIR primer sequences, 5'-AAATATGGCGGCGTCTACACGGA-3'; 5'-TCCAGAACCCTCTGACATTTGCCT-3'; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primer sequences, 5'-GAAGGTGAAGGTCGGAGTC-3'; 5'-GAAGATGGTGATGGGATTTC-3'. The mRNA level was normalized by GAPDH. We use 2 (−ΔΔCt) calculated the results of real-time PCR amplification (8).
MTT assay
Cells were seeded on 96 wells, and then the margins were washed using a phosphate buffered saline (PBS) buffer. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Boster Biotech, Wuhan, Hubei, China) was added at about 0, 24, 48, and 96 h after transfection, followed by replacing with dimethyl sulfoxide (DMSO) 4 h later. The mixture was incubated in an incubator for 5–10 minutes. The absorbance values of the experimental group and the blank control group were determined by enzyme-linked immunosorbent assay (ELISA) at 570 nm. The survival rate was the ratio of the optical density (OD) value between the experimental group and the blank control group. The test was performed at least in triplicate.
Scratch assay
The cells were seeded on 6-well plates, and a perpendicular direction scratch was performed after transfection. Then, the plates were washed using PBS buffer, and then the migration was recorded at 0 h and 48 h, respectively. The images were analyzed using the Image J software (https://imagej.net). The test was performed at least in triplicate.
Transwell assay
About 24 h after transfection, the cells were incubated on the upper Transwell chamber (Corning, Corning, NY, USA) containing no serum. In the lower chamber, there was high glucose Dulbecco’s Modified Eagle Medium (DMEM) containing 10% FBS. In the invasion assay, we put the Matrigel (Corning, Corning, NY, USA) in the upper compartment. The cells were incubated for 24 h in an incubator, followed by cleaning of the upper cells using a cotton bud. The cells migrated to the lower chamber were fixed using paraformaldehyde, and then stained with 0.1% crystal violet for 20 minutes. Subsequently, the cells were washed 3 times using PBS. A total of 5 visual fields were randomly selected under a magnification of 10×.
Western blot analysis
Extract total proteins from cells, and then the protein content was evaluated. The protein was separated by electrophoresis on a 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to a Hybond-P polyvinylidene difluoride (PVDF) membrane. The membrane was blocked in 5% nonfat milk and incubated with Notch-1, Hes-1, Jagged-1, E-cadherin, and vimentin antibodies (1:1,000, CST, Danvers, MA, USA) 4 °C for the night. Then, incubation with the horseradish peroxidase (HRP)-conjugated secondary antibody (1:2,000, CST, Danvers, MA, USA) and leave them at room temperature for 1 hour. After washing with PBS, the bound primary antibody was visualized with the Enhanced Chemiluminescence System (Amersham, Piscataway, NJ, USA) and exposed to film. -The same approach was applied to GAPDH (Solarbio, San Diego, CA, USA) for loading control. The relative density was analyzed with the Image J software.
Statistical analysis
We express all data as the mean plus or minus standard deviation. The statistical software using statistical software SPSS 22.0 (IBM, Chicago, IL, USA) to analysis data. One-factor analysis of variance (ANOVA) was utilized for the multiple group comparison. The Student’s t-test was used for comparison between groups. The software GraphPad Prism 7 (GraphPad, San Diego, CA, USA) was used for the plot. A P value <0.05 was considered statistically significant.
Results
HOTAIR silencing efficiency
As shown in Figure 1, compared with the normal control, HOTAIR expression was substantial decreased in the Tca-8113 group (P<0.01). Similarly, a significant decrease was noticed in the HOTAIR expression in TSCCA (P<0.01), especially the siRNA-3. On this basis, siRNA-3 was selected for subsequent experiments.
Effects of HOTAIR silencing on the proliferation of tongue cancer cells
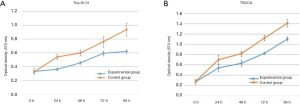
The absorbance in the Tca-8113 cells revealed that there was significant inhibition in the Tca-8113 group compared with that of the control group at 24, 48, 72, and 96 h, respectively (all P<0.01, Figure 2A). In the TSCCA group, significant decline was noticed in the absorbance compared with the control group at 24, 48, 72, and 96 h, respectively (all P<0.01, Figure 2B).
Effects of HOTAIR silencing on the migration of cancer cells
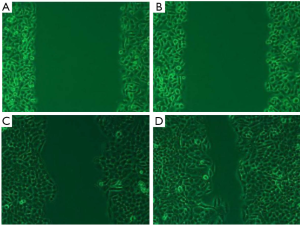
By contrast with the control group, a substantial decline was found in the migration of the cells in the Tca-8113 group (P<0.01, Figure 3). Similarly, significant decline was noticed in the migration of the TSCCA cells compared with the control group (P<0.01). The transwell assay indicated that the number of cells penetrating the small chamber significantly declined contrasted with the control group (P<0.01, Figure 4).

Effects of HOTAIR silencing on the invasion of cancer cells
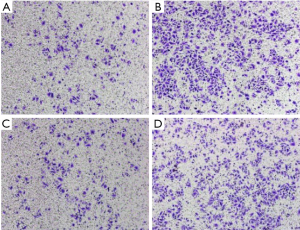
The transwell invasion assay indicated that the number of cells penetrating the membrane in the Tca-8113 cells revealed substantial decline contrasted with that of the control group (230±24.2 vs. 586.4±23.1, P<0.01, Figure 5). In the TSCCA group, the number of cells penetrating the membrane showed significant decline compared with the control group (191.4±17.5 vs. 429.6±26.5, P<0.01, Figure 6). These findings implied that HOTAIR silencing attenuated the invasion of cancer cells.

Expression of EMT related protein after transfection
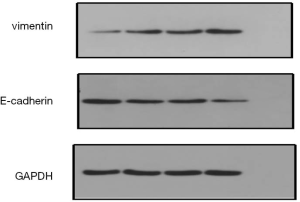
After HOTAIR silencing, there was significant down-regulation in vimentin expression, while the expression of E-cadherin was significant up-regulated (Figure 7). These findings implied that HOTAIR silencing could inhibit the EMT in Tca-8113 and TSCCA cells, while HOTAIR could induce EMT in Tca-8113 and TSCCA cells.
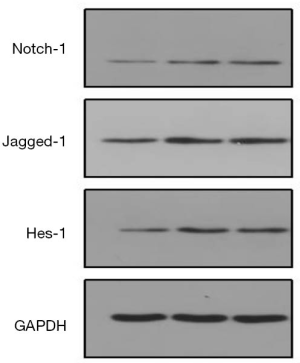
Effects of HOTAIR on the Notch signaling pathway in Tca8113 cells
In the Tca8113 cells, a significant decline was observed in Notch1, Jagged1, and Hes-1 protein expression in the presence of HOTAIR knockdown (P<0.05). Whereas, no statistical differences were discovered in the cells transfected with scramble sequences–contrasted with the blank control (P>0.05, Figure 8).
Discussion
As a novel lncRNA, HOTAIR has been taken a critical role in the regulation of several types of malignancies (9). Nowadays, extensive studies have been focusing on the invasion and metastasis mechanisms of several malignancies (8-12), with the aim of identifying potential targets for treating cancer. HOTAIR is highly expressed in supercutaneous malignancies. HOTAIR regulates target genes through the Hotair5 “structure and 3” structure recruitment protein complex. The high expression of HOTAIR eventually leads to malignant proliferation, reduced apoptosis and distant metastasis of tumor cells, which affects the disease progression of tumor patients and the therapeutic effect of drugs. It has been reported that HOTAIR is aberrantly expressed in several cancers including breast cancer, pancreatic cancer, and esophageal squamous cell carcinoma, which affects the prognosis of these patients (6,8,11-14). In this study, we wanted to explore the effects of lncHOTAIR on the proliferation and migration of Tca-8113 and TSCCA cells. Our data showed that lncHOTAIR could contribute to the invasion and metastasis of these cells.
In vitro experiments have indicated that HOTAIR is involved in the modulation of proliferation, invasion, and apoptosis of cancer cells (6,13,15,16). It has been revealed that HOTAIR is related to the apoptosis, proliferation, invasion, and metastasis of several cancers. For example, after HOTAIR silencing in hepatoma cancer cells, the cellular apoptosis induced by tumor necrosis factor-alpha (TNF-α) was triggered, which obviously attenuated the proliferation, invasion, and metastasis of cancer cells (16). In laryngeal squamous cell carcinoma with knock-down of HOTAIR, the expression of anti-oncogene PTNE was up-regulated, which then led to a block of AKT signaling activation and cell arrest in the G1 phase, as well as inhibiting the cellular proliferation (17). In adenocarcinoma of the uterine cervix (18), HOTAIR could inhibit the expression of p21, which then promoted the cellular proliferation.
In this study, we firstly down-regulated the expression of HOTAIR in the TSCCA and Tca-8113 cells. Then, functional testing showed that HOTAIR contributed to the proliferation, migration, and invasion of these cells. In the MTT assay, HOTAIR knock-down led to a decline of absorbance at each time point compared with the control group, which implied that HOTAIR knock-down attenuated cellular proliferation. In terms of invasion, HOTAIR contributed to the invasion of TSCCA and Tca-8113 cells. Similarly, in the migration assay, the number of cells penetrating the small chamber in the treatment group was substantial higher than that of the control group, indicating that HOTAIR resulted in metastasis of these cells. In the invasion tests, the number of cells penetrating the membrane in the experimental groups was significantly higher than that of the control group, which demonstrated that the invasion capacity declined after HOTAIR silencing.
The process of EMT is the premise for the migration and invasion of cancer cells (19). Several factors and signaling pathways could be involved in the EMT process through corresponding receptor signals. In this study, HOTAIR knock-out led to significant decline in the proliferation, metastasis, and invasion of the TSCCA and Tca-8113 cells, which demonstrated that HOTAIR was associated with the pathogenesis, invasion, and migration of TSCCA. After HOTAIR silencing, the expression of E-cadherin was significantly upregulated and vimentin was down-regulated in the cells. This implied that HOTAIR may induce EMT in Tca-8113 and TSCCA cells. Silencing HOTAIR could reverse the EMT phenotype, inhibit EMT, and then inhibit the invasion and metastasis of cancer.
Current understanding supports that EMT occurs in the early stage of invasion of malignant cancer cells. To our best knowledge, Notch signaling is crucial for EMT induced by hypoxia, invasion, and metastasis (19). As a highly conserved signaling pathway, the Notch signaling pathway exists in nearly all organisms, and it is involved in cellular proliferation, differentiation, and apoptosis. Moreover, it participates in embryonic development, T lymphocyte development, self-renewal of the hematopoietic stem cells, and also regulates angiogenesis (20). Notch signaling pathway molecules have been aberrantly expressed in several malignancies; for instance, Notch-1 receptor and down-stream target gene (Hes-10) overexpression has been associated with staging and drug tolerance of malignant cancers. Notch-3 receptor expression has been associated with the relapse of malignant cancer. Moreover, several studies have demonstrated that the Notch signaling pathway interacted with lncRNAs. For example, Notch-1 induced up-regulation of lncRNA GAS5 and contributed to the metastasis of breast cancer cells (21), while lncRNA NALT up-regulated Notch signaling pathway modulated the pathogenesis of acute lymphocytic leukemia (22). In a previous study, Dong et al. (9) reported that HOTAIR was upregulated in retinoblastoma compared to the adjacent cancer tissues, and the Notch signaling pathway in the retinoblastoma was aberrantly activated. Meanwhile, HOTAIR knock-down resulted in attenuation of the Notch signaling pathway, which implied that HOTAIR could regulate the Notch signaling pathway. In this study, our data revealed that Notch1, Jagged 1, and Hes-1 were down-regulated in the TSCCA after HOTAIR knock-down, which then inhibited the growth of cancer cells.
Niland, Merry, and Khalil showed that HOTAIR was closely related to the invasion, metastasis, and EMT of TSCCA (23). The Notch signaling pathway involved in the proliferation, differentiation, and apoptosis was closely related to HOTAIR. Our study showed that HOTAIR could contribute to the invasion and migration of Tca-8113 and TSCCA, which was closely related to EMT. Knock-down of HOTAIR could attenuate the expression of Jagged-1, Notch-1 receptor, and Hes-1, which then inhibited the proliferation of Tca-8113. We speculated that HOTAIR may serve as a regulator for the cellular proliferation, differentiation, and apoptosis of cancer cells. This demonstrated that HOTAIR modulated the Notch signaling pathway involved in the pathogenesis of tongue cancer.
There were some limitations in this study. The roles of the Notch signaling pathway are various in different cancer types at different stages. Additionally, monitoring the Notch signaling pathway in a dynamic manner remains challenging.
In conclusion, HOTAIR and the Notch signaling pathway are associated with the invasion and metastasis of tongue cancer. The Notch signaling pathway was positively correlated with the HOTAIR signaling pathway, which implied that HOTAIR could modulate the Notch signaling pathway. In future, further studies are required to investigate the mutual interactions of HOTAIR and the Notch signaling pathway.
Acknowledgments
Funding: The study was supported by
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tcr-20-3452
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-3452
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-3452). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocols were approved by the Ethical Committee of The First Affiliated Hospital of Xinjiang Medical University (No. K202012-05).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pulte D, Brenner H. Changes in Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncologist 2010;15:994-1001. [Crossref] [PubMed]
- Warnakulasuriya S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol 2009;45:309-16. [Crossref] [PubMed]
- Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007;316:1484-8. [Crossref] [PubMed]
- Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog 2013;52:908-15. [Crossref] [PubMed]
- Fan X, Huang H, Ji Z, Mao Q. Long non-coding RNA MEG3 functions as a competing endogenous RNA of miR-93 to regulate bladder cancer progression via PI3K/AKT/mTOR pathway. Transl Cancer Res 2020;9:1678-88. [Crossref]
- Jia X, Meng W, Zhang L, et al. Construction of differentially expressed Her-2 related lncRNA-mRNA-miRNA ceRNA network in Her-2 positive breast cancer. Transl Cancer Res 2020;9:2527-33. [Crossref]
- Wu Y, Zhang L, Zhang L, et al. Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol 2015;46:2586-94. [Crossref] [PubMed]
- Tang L, Zhang W, Su B, et al. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int 2013;2013:251098. [Crossref] [PubMed]
- Dong C, Liu S, Lv Y, et al. Long non-coding RNA HOTAIR regulates proliferation and invasion via activating Notch signalling pathway in retinoblastoma. J Biosci 2016;41:677-87. [Crossref] [PubMed]
- Nie Y, Liu X, Qu S, et al. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013;104:458-64. [Crossref] [PubMed]
- Sørensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat 2013;142:529-36. [Crossref] [PubMed]
- Sun L, Fang J. Writer meets eraser in HOTAIR. Acta Biochim Biophys Sin (Shanghai) 2011;43:1-3. [Crossref] [PubMed]
- Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011;71:6320-6. [Crossref] [PubMed]
- Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013;13:464. [Crossref] [PubMed]
- Ono H, Motoi N, Nagano H, et al. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med 2014;3:632-42. [Crossref] [PubMed]
- Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol 2014;134:121-8. [Crossref] [PubMed]
- Lee M, Kim HJ, Kim SW, et al. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget 2016;7:44558-71. [Crossref] [PubMed]
- Bhan A, Hussain I, Ansari KI, et al. Bisphenol-A and diethylstilbestrol exposure induces the expression of breast cancer associated long noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol Biol 2014;141:160-70.
- Ishida T, Hijioka H, Kume K, et al. Notch signaling induces EMT in OSCC cell lines in a hypoxic environment. Oncol Lett 2013;6:1201-6. [Crossref] [PubMed]
- Leong K G.. Recent insights into the role of Notch signaling in tumorigenesis. Blood 2006;107:2223-33. [Crossref] [PubMed]
- Pei J, Wang B. Notch-1 promotes breast cancer cells proliferation by regulating LncRNA GAS5. Int J Clin Exp Med 2015;8:14464-71. [PubMed]
- Wang Y, Wu P, Lin R, et al. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci Rep 2015;5:13749. [Crossref] [PubMed]
- Niland CN, Merry CR, Khalil AM. Emerging Roles for Long Non-Coding RNAs in Cancer and Neurological Disorders. Front Genet 2012;3:25. [Crossref] [PubMed]
(English Language Editor: J. Jones)



