
A rare multiple primary sarcomatoid carcinoma (SCA) of small intestine harboring driver gene mutations: a case report and a literature review
Introduction
Primary sarcomatoid carcinoma (SCA) is a type of rare tumor consisting of both malignant epithelial and mesenchymal components (1). Tumors with sarcomatoid features have been reported to be more aggressive (2), while the pathogenesis of SCA has not been elucidated. Collision theory is a popular hypothesis suggesting that two different types of tumor cells originate from mesenchymal and epithelial origins separately (3). However, a more reasonable theory that sarcomatoid and carcinomatoid elements sharing a common clonal origin is supported by recent studies based on genomic sequencing (4,5). SCA has been reported in various organs, including lung, uterine, salivary and thyroid glands (6,7). In small intestine, SCA was described using the term enteroblastoma for the first time in 1973 (8), and other terms such as SCA, carcinosarcoma, metaplastic carcinoma, and spindle cell carcinoma, were subsequently used in other organs. Nowadays, SCA is the most accepted term used in diagnostic surgical reports (9). SCA can be discriminated from polyps by pathological examinations. SCA has both epithelioid components and sarcomatoid components with high dysplasia, and positive staining of NSE, CK and vimentin can be observed by immunohistochemistry. These features cannot be found in polyps, which are featured by hyperplasia with generally normal adenoid structure.
The most frequent types of SCA, including pulmonary SCA, sarcomatoid renal cell carcinoma, and uterine carcinosarcoma, have been characterized in terms of diagnostic classification and molecular mechanism (10-12), while intestinal SCAs are very unusual. Due to inaccessibility of routine endoscopy and nonspecific clinical symptoms, patients affected by SCA were usually diagnosed at late stages. Only dozens of cases were reported (13,14) and the patients generally had poor prognosis. We herein report a male with multiple (over 100) primary jejunum SCAs scattered along the whole jejunum and proximal ileum, which has never been reported in previous SCA studies. We also established the whole-exome mutational profile of SCA for the first time, and identified featured SNV/INDEL and CNV alterations, and revealed key tumor driver gene mutations and aberrant functional pathways. We present the following article in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2829).
Case presentation
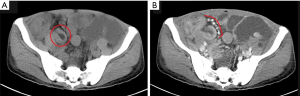
A 54-year-old Chinese male presented with abdominal distension, fatigue and loss of weight and was diagnosed with gastro and duodenal inflammation by gastroscopy with anemia at a local hospital. Abdomen ultrasonic examination was performed with no signs of abnormality. He was referred to our hospital due to symptoms aggravated within two weeks. Preoperative contrast-enhanced computed tomography (CT) showed multiple polypoid lesions in small intestine causing intussusceptions and obstruction (Figure 1A,B). No masses were seen in lung, liver, or pancreas. Laparotomy was then performed and approximately 1,000 mL ascites in the peritoneal cavity were found. Meanwhile, many polypoid lumps in small bowel were observed with enlarged regional lymph nodes. No lesion was found in other parts of the gastrointestinal tract. Segmental resection of his whole jejunum and proximal ileum (total length of 300 cm, distal resected margin at 160 cm to the ileocecal valve) along with seven mesentery lymph nodes were performed.
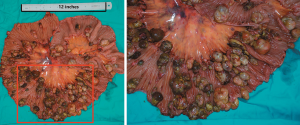
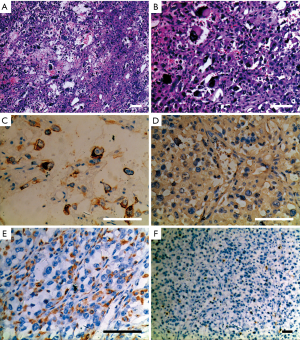
More than 100 round-like polypoid masses with diameter from 1.5 to 2.6 cm were dispersing along the resected intestinal lumen (Figure 2). Metastases were found in all resected lymph nodes. Microscopically, the tumor was composed of two different components of cells, the pleomorphic cells with giant nuclei and the epithelioid cells. The two components were present in complex form without clear separation, in which approximately 30% of the lesions belonged to epithelioid components and 70% belonged to sarcomatoid components. Immunohistochemical staining showed vimentin(+), CK(+), CK8(+), CK18(+), CD34(+), CD68(+), S-100(-), Dog-1(-), CD117(-), CD3(-), CD20(-), CD30(-), CD57(-), desmin(-), CyclinD1(-), and SMA(-), suggesting both epithelial and stromal components (Figure 3). The final diagnosis was confirmed as jejunal SCA with mesenteric lymph nodes metastasis, pT3N2M0, stage IV. The patient died 2 months after surgery due to severe malnutrition, cachexia and electrolyte disturbance following one cycle of postoperative chemotherapy (IFO+Epirubicin).
The tumor and its adjacent normal tissue were fixed with formalin and embedded with paraffin (FFPE). To further investigate the genomic features of this tumor, whole-exome sequencing was performed with DNA extracted from both FFPE samples. The purity and concentration of the DNA fragments were assessed using the Qubit 2.0 fluorometer and the Qubit. DNA sequencing was then performed on the Illumina Novaseq6000 system according to the manufacturer’s recommendations at an average depth of 5,000×. Sequencing data were de-multiplexed and aligned to the human reference genome (hg19 or GRch37) using Burrows-Wheeler Aligner (version 0.7.15)-r1140 by default settings. Pileup files for properly paired reads with mapping quality ≥60 was generated using Samtools (http://www.htslib.org/). Thirty-five germline alterations were identified from normal tissue using a 58-gene analysis pipeline. According to the latest American College of Medical Genetics and Genomics (ACMG) guidelines, none was interpreted as pathogenic and only 3 as variant of undetermined significance (VUS) (Table S1).
Somatic variants lists were created using VarScan2 (http://varscan.sourceforge.net/). Allele frequencies were calculated for all Q30 bases. Using a custom Python script, previously identified tumor DNA mutations were intersected with a Samtools pileup file generated for each sample, and the number and frequency were then calculated for each mutation. A mutation was identified if ≥5 mutant reads were identified and ≥1 mutant read was identified on each strand. Two hundred and seventy-six single nucleotide variants (SNVs) (Table S2), 8 short deletions and 2 short insertions (Table S3) were identified in the tumor tissue, including 38 point and indel alterations in driver genes defined by previous studies (15-18) (Table 1). Sixty-nine copy number variations were also detected (Table S4), mainly gathered in chromosome 6, 7, 16 and 20 (Figure 4). The tumor mutation burden (TMB) was 7.15 mutations/Mb. Several key driver genes were revealed to harbor mutations, including KRAS (c.37G>T, 66.3%), TP53 (c.871A>T, 47.7%), EGFR (c.1351C>T, 4.2%), CDKN2A (c.104_138del, 11.1%). No alteration was found in PDGFR gene, which is usually mutated in GIST.
Table 1
| ID | Gene | Exon | Nucleotide | Protein | Allele frequency | Variant type |
|---|---|---|---|---|---|---|
| 1 | AARS2 | Exon14 | c.G1961T | p.G654V | 31.26 | Snv |
| 2 | ATF7IP | Exon2 | c.C1154T | p.A385V | 6.52 | Snv |
| 3 | ATP2B3 | Exon14 | c.G2396T | p.G799V | 66.67 | Snv |
| 4 | BAZ2A | Exon9 | c.C1795T | p.R599C | 10.37 | Snv |
| 5 | BIRC6 | Exon10 | c.G2848C | p.D950H | 28.82 | Snv |
| 6 | CDKN2A | Exon1 | c.104_138del | p.G35Efs*73 | 11.11 | Indel |
| 7 | CEP170 | Exon13 | c.C2375A | p.S792X | 11.67 | Snv |
| 8 | CFH | Exon8 | c.C1126A | p.Q376K | 11.83 | Snv |
| 9 | CREB3L1 | Exon3 | c.C461T | p.A154V | 5.94 | Snv |
| 10 | CSF3R | Exon13 | c.C1655A | p.P552H | 16.74 | Snv |
| 11 | DST | Exon24 | c.A6151G | p.R2051G | 38.77 | Snv |
| 12 | ECT2L | Exon8 | c.884delC | p.R296Gfs*8 | 50.85 | Indel |
| 13 | EGFR | Exon12 | c.C1351T | p.R451C | 4.21 | Snv |
| 14 | EPB41L3 | Exon12 | c.A1355T | p.Q452L | 12.89 | Snv |
| 15 | EPHA7 | Exon1 | c.G85T | p.A29S | 58.72 | Snv |
| 16 | FAT3 | Exon23 | c.G12328C | p.G4110R | 19.95 | Snv |
| 17 | FBN2 | Exon38 | c.C4892A | p.T1631N | 55.56 | Snv |
| 18 | GNAS | Exon1 | c.C1336T | p.P446S | 10.71 | Snv |
| 19 | GRIN2A | Intron12 | c.2356+1G>A | nil | 29.08 | Snv |
| 20 | IRS4 | Exon1 | c.G1982A | p.R661K | 60.69 | Snv |
| 21 | ITGA6 | Exon13 | c.C1786T | p.R596X | 14.62 | Snv |
| 22 | KRAS | Exon2 | c.G37T | p.G13C | 66.26 | Snv |
| 23 | LRP1B | Exon67 | c.G10481T | p.R3494L | 21.88 | Snv |
| 24 | MAPK8IP1 | Exon8 | c.A1697T | p.Q566L | 42.38 | Snv |
| 25 | MAST2 | Exon1 | c.C19T | p.R7C | 26.09 | Snv |
| 26 | MKL1 | Exon12 | c.C1853T | p.P618L | 5.04 | Snv |
| 27 | NAV3 | Exon1 | c.C170T | p.A57V | 45.74 | Snv |
| 28 | NAV3 | Exon5 | c.C539T | p.S180F | 15.88 | Snv |
| 29 | PDGFB | Exon4 | c.G268T | p.E90X | 48.46 | Snv |
| 30 | POT1 | Exon7 | c.G248T | p.R83M | 59.35 | Snv |
| 31 | SMARCD1 | Intron11 | c.1393-1G>A | nil | 16.19 | Snv |
| 32 | TP53 | Exon8 | c.A871T | p.K291X | 47.72 | Snv |
| 33 | TSHZ2 | Exon2 | c.T1763C | p.V588A | 18.81 | Snv |
| 34 | USP8 | Exon15 | c.C2287T | p.R763W | 14.19 | Snv |
| 35 | USP8 | Exon15 | c.C2292A | p.N764K | 15.44 | Snv |
| 36 | USP9X | Exon26 | c.G3920A | p.S1307N | 5.36 | Snv |
| 37 | ZBTB16 | Exon2 | c.G1174T | p.A392S | 33.64 | Snv |
| 38 | ZNRF3 | Exon8 | c.G2380T | p.G794C | 18.75 | Snv |
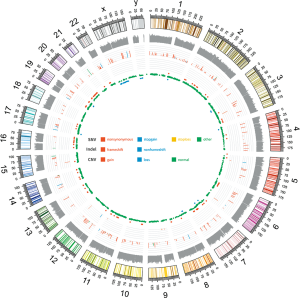
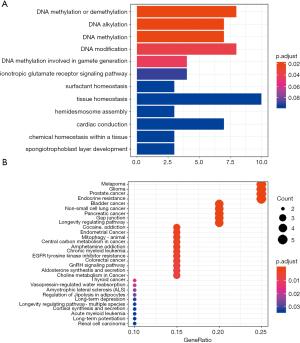
Functional clustering analysis was employed on somatic mutations. Using clusterProfiler (19), we found most enriched GO term was DNA methylation or demethylation. KEGG clustering analysis (BH-corrected, P<0.05) showed several cancer-related pathways (Figure 5). These observations suggest that the genetic abnormalities in this case were distinct from other SCA cases, and reflected the uniqueness of this case.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patients.
Discussion and conclusions
Small bowel tumors are not commonly seen, accounting for less than 5% of all gastrointestinal cancers. The most common type of small bowel malignancies is adenocarcinoma, followed by neuroendocrine tumor, stromal tumor, and lymphoma (20,21). SCA is very rare. Only 32 cases of SCA in the small bowel have been reported in the literature to date (Table 2). The tumor mainly occurs in elder patients with a mean age of 60 years old (ranged from 35 to 85, Figure 6A) and a male-female ratio of 1.46:1 (19 male vs. 13 female). The most frequent primary location is jejunum (17/32, 53.1%) followed by ileum (14/32, 43.8%), and duodenum SCA is very rare (1/32, 3.1%). Mesenteric lymph nodes metastasis was present in 56.3% (18/32) of the reported cases. Macroscopically, SCA can be divided into five types, including the endophytic (33.3%), the polypoid (29.6%), the ulcerating (18.5%), the nodular (11.1%) and the exophytic (7.4%) (Table 2, Figure 6B). The case in this study belonged to the polypoid type. Microscopically, SCA tumors are composed of two or three cells components: polygonal, anaplastic and spindle, and basically exhibited positive expression for both cytokeratin (CK) (27 positives in 30 patients) and vimentin (21 positives in 21 patients) in immunohistochemistry staining (Table 2). Most patients had surgical resection but only survived for several months. The median overall survival (OS) was 7 months (Table 2, Figure 6C).
Table 2
| ID | Age | Gender | Diagnosis | Tumor Site | No of lesion(s) | Maximal Diameter (cm) | Morphology | Metastasis | CK | Vimentin | OS (months) | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 44 | M | Enteroblastoma | Ileum | 1 | N/A | Polypoid | Yes | N/A | N/A | N/A | (22) |
| 2 | 35 | F | Anasplastic and SCA | Jejunum | 1 | 7.5 | Endophytic | Yes | − | N/A | 36 | (2) |
| 3 | 38 | F | Anasplastic and SCA | Jejunum | 1 | 16 | Endophytic | Yes | + | N/A | 8 | (2) |
| 4 | 48 | F | Anasplastic and SCA | Jejunum | 1 | 6 | Endophytic | Yes | + | N/A | 29 | (2) |
| 5 | 65 | M | Anasplastic and SCA | Jejunum | 1 | 5 | Endophytic | Yes | + | N/A | 5 | (2) |
| 6 | 54 | F | Anasplastic and SCA | Ileum | 1 | 4.5 | Endophytic | No | − | N/A | 12* | (2) |
| 7 | 62 | M | Anasplastic and SCA | Ileum | 1 | 5 | Endophytic | Yes | − | N/A | 20 | (2) |
| 8 | 52 | F | Pleomorphic CA | Jejunum | 2 | 8 | Nodular | Yes | + | + | 7 | (23) |
| 9 | 56 | M | Pleomorphic CA | Jejunum | 2 | 8 | Nodular | Yes | + | + | 8 | (23) |
| 10 | 45 | M | Pleomorphic CA | Ileum | 1 | 3 | Endophytic | No | + | + | 0.2 | (24) |
| 11 | 57 | M | Pleomorphic CA | Ileum | 1 | 14 | Endophytic | No | + | + | 6* | (24) |
| 12 | 63 | M | Pleomorphic CA | Ileum | 1 | 6 | Endophytic | No | + | + | 39* | (24) |
| 13 | 68 | F | SCA | Ileum | 1 | N/A | N/A | No | N/A | N/A | N/A | (25) |
| 14 | 75 | M | SCA | Ileum | 1 | N/A | N/A | No | + | + | N/A | (25) |
| 15 | 77 | M | SCA | Duodenum | 1 | N/A | N/A | Yes | + | + | N/A | (25) |
| 16 | 76 | F | SCA | Jejunum | N/A | N/A | N/A | No | + | + | 2 | (26) |
| 17 | 76 | F | SCA | Ileum | 1 | 5 | Ulcerating | NA | + | + | 2 | (27) |
| 18 | 53 | M | Anasplastic and SCA | Ileum | N/A | N/A | Polypoid | Yes | + | + | N/A | (28) |
| 19 | 56 | M | SCA | Ileum | 1 | 9.2 | Ulcerating | Yes | + | + | 3 | (29) |
| 20 | 55 | M | SCA | Jejunum | 1 | 7.5 | Polypoid | Yes | + | + | 11 | (1) |
| 21 | 55 | M | SCA | Jejunum | N/A | N/A | N/A | Yes | + | N/A | 9.4 | (30) |
| 22 | 51 | F | SCA | Jejunum | 1 | 8 | Polypoid | Yes | + | + | 1.9 | (31) |
| 23 | 85 | F | SCA | Jejunum | 1 | 10.1 | Polypoid | No | + | N/A | 3 | (32) |
| 24 | 70 | F | SCA | Jejunum | 1 | NA | Polypoid | No | + | + | 7* | (33) |
| 25 | 56 | F | SCA | Jejunum | 1 | 6.7 | Ulcerating | Yes | + | + | 6 | (34) |
| 26 | 62 | M | SCA | Ileum | 1 | 15 | Ulcerating | No | + | + | 3* | (35) |
| 27 | 69 | M | N/A | Jejunum | 1 | 6 | Polypoid | No | + | + | 41* | (36) |
| 28 | 78 | M | SCA | Jejunum | N/A | N/A | Exophytic | NA | + | + | N/A | (37) |
| 29 | 60 | M | N/A | Ileum | N/A | N/A | Nodular | Yes | + | N/A | N/A | (38) |
| 30 | 60 | M | SCA | Jejunum | 6 | 5 | Ulcerating | Yes | + | + | 0.33 | (17) |
| 31 | 62 | M | SCA | Jejunum | 1 | 12 | Exophytic | Yes | + | + | 1 | (39) |
| 32 | 58 | F | SCA | Ileum | 1 | 3 | Polypoid | No | + | + | 0.36 | (15) |
| This study | 54 | M | SCA | Jejunum | >100 | 2.6 | Polypoid | No | + | + | 3 | This study |

In our case, aggressive development was observed following the appearance of symptoms in gastrointestinal tract. Tumor location identification was difficult and ambiguous. A very distinct clinical feature of our case is that many lesions scattered in jejunum and proximal ileum. Compare to the single, large tumors (average diameter was 7.75 cm) in most reported cases (Table 2, Figure 6D), our patient had multiple smaller tumors (1.5 to 2.6 cm), which is characteristic from those previous reported.
Immunohistochemistry, in combination with H&E staining, is the golden standard for diagnosis of SCA. A wide panel of markers has been used for SCA pathological diagnosis. SCA usually presents positive for CK, vimentin, EMA, and negative for desmin, S-100, and DOG-1. C-kit negativity is the key to differentiate SCA from GIST, which has similar morphology with SCA (9,40). Certain cases may also exhibit focal positivity for neuroendocrine and neuron-specific markers (1).
Exploration in genetic alterations of small intestine SCA had not been conducted. We exploited next generation sequencing (NGS) technique to study the whole-exome genetic profile of this case. Among 35 germline alterations, none was interpreted as known pathogenic mutation and only 3 were interpreted VUS according to ACMG guidelines. This might suggest the carcinogenesis of the tumor in our case was driven by some acquired factors.
In TCGA data and other large-scale analysis of various types of sarcoma, the top frequently mutated genes include TP53, TTN, ATRX, PIK3CA, MUC16, RB1, and PTEN (12,41,42). PI3K signal pathway is undoubtedly a hotspot pathway in this disease based on previous studies. Aberrances on driver genes in this pathway are involved in the progression of cancer. However, the mutated profile of our case did not show that PI3K signal pathway was the dominant abnormality. We identified cetuximab-resistant mutation in KRAS gene (c.37G>T, p.G13C), which is in upstream of PI3K signal pathway. This mutation could lead to activation of the downstream signal pathways (12,41-43). The specific alteration in TP53 (c.871A>T, p.K291*) is only described in a few cancer studies, including those on transitional cell (urothelial) carcinoma (44), large intestine adenocarcinoma (22), laryngeal squamous cell carcinoma (23), and melanoma (45). CDKN2A gene encodes tumor suppressor proteins which act as negative regulator in the proliferation of normal cells and induce cell cycle arrest in G1 and G2 phases. The CDKN2A mutation (c.104_138del, p.G35fs) is a frameshift mutation which could lead to malfunctioned truncated protein. Many amplified genes were found in our study, but their roles were not clarified. It is possible that the combination of multiple aberrances in key driver genes with other genetic alterations led to the characteristics of the tumor, but the key factors in its pathogenesis still needs further investigation.
Small intestinal cancers mainly include adenocarcinoma, carcinoid, malignant lymphoma and sarcoma, which account for 2–3% of all gastrointestinal cancers. It was reported that 55–80% of them are adenocarcinoma and carcinoid, while lymphoma and sarcoma are rarely seen (46,47). The mechanism of small intestine adenocarcinoma has been suggested to be similar to that of the colorectal cancer, including APC, TP53 and KRAS mutations, aberrancies of the Wnt pathway and abnormal mismatch repair (48). The mechanism of carcinoid was suggested to be related to TGF-β pathway (49) and Chromosome X inactivation (50). SCA is the rarest type of small intestine carcinoma, and most reports so far are case reports without systematic investigation on its molecular mechanism. Our study provided the first piece of evidence on the possible molecular mechanism of small intestine SCA.
There is still no official treatment guideline for SCA. Palliative segment resection was the main treatment in most cases. Adjuvant chemotherapy, such as 5-FU and/or cisplatin or radiotherapy, was performed in some patients, but no report identified improvements in survival. In conclusion, diagnosis and treatment of SCA are still clinical challenges. Our sequencing results revealed the genomic feature of a rare SCA case, providing further understanding on molecular pathogenesis of this specific cancer.
Acknowledgments
The authors thank the patient and patient’s kin for agreement to publication of the report. We appreciate Dr. Dibesh Shrestha for proof reading and checking the English language of the manuscript as a native speaker.
Funding: This study was supported by the grants from
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2829
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2829). Dr. Zhu, Dr. Li, Dr. Wen, Dr. Ji, Dr. Ruize Zhou, Dr. Tuo and Dr. Huang report grants from Project of Yunnan Science and Technology Department, grants from Yunnan Provincial Health Commission Medical Reserve Talents Training Program, during the conduct of the study. Dr. Lele Song reports grants from Yunnan Provincial Health Commission Medical Reserve Talents Training Program, grants from Project of Yunnan Science and Technology Department, grants from the Special Funds for Strategic Emerging Industry Development of Shenzhen, grants from and the Science and Technology Project of Shenzhen, during the conduct of the study; reports other from HaploX Biotechnology, Shenzhen, China, outside the submitted work. The other authors report grants from the Special Funds for Strategic Emerging Industry Development of Shenzhen, grants from and the Science and Technology Project of Shenzhen, during the conduct of the study; report other from HaploX Biotechnology, Shenzhen, China, outside the submitted work.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Reid-Nicholson M, Idrees M, Perino G, et al. Sarcomatoid carcinoma of the small intestine: a case report and review of the literature. Arch Pathol Lab Med 2004;128:918-21. [Crossref] [PubMed]
- Robey-Cafferty SS, Silva EG, Cleary KR. Anaplastic and sarcomatoid carcinoma of the small intestine: a clinico-pathologic study. Hum Pathol 1989;20:858-63. [Crossref] [PubMed]
- Holtz F, Fox JE, Abell MR. Carcinosarcoma of the urinary bladder. Cancer-Am Cancer Soc 1972;29:294-304. [PubMed]
- Armstrong AB, Wang M, Eble JN, et al. TP53 mutational analysis supports monoclonal origin of biphasic sarcomatoid urothelial carcinoma (carcinosarcoma) of the urinary bladder. Mod Pathol 2009;22:113-8. [Crossref] [PubMed]
- Bi M, Zhao S, Said JW, et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proc Natl Acad Sci U S A 2016;113:2170-5. [Crossref] [PubMed]
- Iezzoni JC, Mills SE. Sarcomatoid carcinomas (carcinosarcomas) of the gastrointestinal tract: a review. Semin Diagn Pathol 1993;10:176-87. [PubMed]
- Guarino M, Tricomi P, Giordano F, et al. Sarcomatoid carcinomas: pathological and histopathogenetic considerations. Pathology 1996;28:298-305. [Crossref] [PubMed]
- Dikman SH, Toker C. Enteroblastoma complicating regional enteritis. Gastroenterology 1973;65:462-6. [Crossref] [PubMed]
- Kwok CM. Sarcomatoid carcinoma of the jejunum with gastric metastases: A case report and review of the literature. Int J Surg Case Rep 2016;28:161-4. [Crossref] [PubMed]
- Liu X, Jia Y, Stoopler MB, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol 2016;34:794-802. [Crossref] [PubMed]
- Wang Z, Kim TB, Peng B, et al. Sarcomatoid Renal Cell Carcinoma Has a Distinct Molecular Pathogenesis, Driver Mutation Profile, and Transcriptional Landscape. Clin Cancer Res 2017;23:6686-96. [Crossref] [PubMed]
- Cherniack AD, Shen H, Walter V, et al. Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell 2017;31:411-23. [Crossref] [PubMed]
- Shim HJ, Hong YK, Kim SJ, et al. Carcinosarcoma on ascending colon found by bowel perforation: a case report. J Korean Soc Coloproctol 2010;26:368-72. [Crossref] [PubMed]
- Arao Y, Kamimura K, Ikemi M, et al. A Rare Duodenal Carcinosarcoma: A Case Report and Literature Review. Intern Med 2019;58:1273-8. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333-9. [Crossref] [PubMed]
- Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep 2013;3:2650. [Crossref] [PubMed]
- Sondka Z, Bamford S, Cole CG, et al. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696-705. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. [Crossref] [PubMed]
- Sarosiek T, Stelmaszuk M. (Small intestine neoplasms). Pol Merkur Lekarski 2018;44:45-8. [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015;65:5-29. [Crossref] [PubMed]
- Nyiraneza C, Jouret-Mourin A, Kartheuser A, et al. Distinctive patterns of p53 protein expression and microsatellite instability in human colorectal cancer. Hum Pathol 2011;42:1897-910. [Crossref] [PubMed]
- Todorova TA, Jordanov SH, Stancheva GS, et al. Mutational Status of CDKN2A and TP53 Genes in Laryngeal Squamous Cell Carcinoma. Pathol Oncol Res 2015;21:413-21. [Crossref] [PubMed]
- Bak M, Teglbjaerg PS. Pleomorphic (giant cell) carcinoma of the intestine. An immunohistochemical and electron microscopic study. Cancer-Am Cancer Soc 1989;64:2557-64. [Crossref] [PubMed]
- Jones EA, Flejou JF, Molas G, et al. Pleomorphic carcinoma of the small bowel. The limitations of immunohistochemical specificity. Pathol Res Pract 1991;187:235-40. [Crossref] [PubMed]
- Fukuda T, Kamishima T, Ohnishi Y, et al. Sarcomatoid carcinoma of the small intestine: histologic, immunohistochemical and ultrastructural features of three cases and its differential diagnosis. Pathol Int 1996;46:682-8. [Crossref] [PubMed]
- Lam KY, Leung CY, Ho JW. Sarcomatoid carcinoma of the small intestine. Aust N Z J Surg 1996;66:636-9. [Crossref] [PubMed]
- Cavazza A, Sandona F, Gelli MC, et al. Monophasic sarcomatoid carcinoma of the ileum. Description of a case and review of the literature. Pathologica 1997;89:425-31. [PubMed]
- Agrawal S, Trivedi MH, Lukens FJ, et al. Anaplastic and sarcomatoid carcinoma of the small intestine: an unusual tumor. J Clin Gastroenterol 1999;29:99-101. [Crossref] [PubMed]
- Tsukadaira A, Koizumi T, Okubo Y, et al. Small-intestinal sarcomatoid carcinoma with superior vena cava syndrome. J Gastroenterol 2002;37:471-5. [Crossref] [PubMed]
- Moriwaki Y, Sugiyama M. Severe anemia inducing preshock caused by sarcomatoid carcinoma of the small intestine. Int Surg 2009;94:164-70. [PubMed]
- Yucel AF, Kocakusak A, Arikan S, et al. A rare cause of acute abdomen: perforated primary sarcomatoid carcinoma of the small intestine - report of a case, with a brief review of the literature. J Cancer Res Ther 2011;7:348-50. [Crossref] [PubMed]
- Pata F, Sengodan M, Tang CB, et al. Concomitant jejunal sarcomatoid carcinoma and gastric GIST in patient with polymyalgia rheumatica: A case report. Int J Surg Case Rep 2013;4:449-52. [Crossref] [PubMed]
- Han N, Han QH, Liu YZ, et al. Perforated sarcomatoid carcinoma of the jejunum: Case report. Oncol Lett 2013;6:562-4. [Crossref] [PubMed]
- Alfonso PN, Jimenez-Alfaro LC, Garcia HM. Sarcomatoid carcinoma of the jejunum presenting as obscure gastrointestinal bleeding in a patient with a history of gliosarcoma. Gastroenterol Rep (Oxf) 2014;2:150-3. [Crossref] [PubMed]
- Lee SE, Park SY. Sarcomatoid carcinoma of the small intestine: a rare and highly aggressive tumor. J Korean Surg Soc 2012;83:321-4. [Crossref] [PubMed]
- Padma SK, Permi HS, Patil CN, et al. Aggressive monophasic sarcomatoid carcinoma of small intestine—a rare case report with review of literature. NUJHS 2012;2:45-7. [Crossref]
- Mittal M, Parikh DA, Jess H, et al. First Case of Small Bowel Sarcomatoid Carcinoma Found by Video Capsule Endoscopy. Gastroenterology Res 2012;5:71-3. [Crossref] [PubMed]
- Kang KA, Yoon JH, Kang G. Sarcomatoid carcinoma of the small intestine: a case report and review of the literature. J Korean Soc Radiol 2013;69:295-9. [Crossref]
- Lee HM, Cho MS, Kim YI. A Surgically Resected Large Sarcomatoid Carcinoma of the Jejunum: A Case Report and Literature Review. J Gastric Cancer 2015;15:143-6. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidi-mensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Ogura K, Hosoda F, Arai Y, et al. Integrated genetic and epigenetic analysis of myxofibrosarcoma. Nat Commun 2018;9:2765. [Crossref] [PubMed]
- Stolze B, Reinhart S, Bulllinger L, et al. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep 2015;5:8535. [Crossref] [PubMed]
- Grollman AP, Shibutani S, Moriya M, et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc Natl Acad Sci U S A 2007;104:12129-34. [Crossref] [PubMed]
- Siroy AE, Boland GM, Milton DR, et al. Beyond BRAF(V600): clinical mutation panel testing by next-generation sequencing in advanced melanoma. J Invest Dermatol 2015;135:508-15. [Crossref] [PubMed]
- Gill SS, Heuman DM, Mihas AA. Small intestinal neoplasms. J Clin Gastroenterol 2001;33:267-82. [Crossref] [PubMed]
- Neugut AI, Marvin MR, Rella VA, et al. An overview of adenocarcinoma of the small intestine. Oncology (Williston Park) 1997;11:529-50. [PubMed]
- Zhang MQ, Chen ZM, Wang HL. Immunohistochemical investigation of tumorigenic pathways in small intestinal adenocarcinoma: a comparison with colorectal adenocarcinoma. Mod Pathol 2006;19:573-80. [Crossref] [PubMed]
- Kidd M, Eick G, Shapiro MD, et al. Microsatellite instability and gene mutations in transforming growth factor-beta type II receptor are absent in small bowel carcinoid tumors. Cancer 2005;103:229-36. [Crossref] [PubMed]
- Guo Z, Li Q, Wilander E, et al. Clonality analysis of multifocal carcinoid tumours of the small intestine by X-chromosome inactivation analysis. J Pathol 2000;190:76-9. [Crossref] [PubMed]


