
KRAS, YWHAE, SP1 and MSRA as biomarkers in endometrial cancer
Introduction
Endometrial cancer (EC) is the most common gynecologic cancer in the United States, with the American Cancer Society expecting that there will be 65,620 new cases in 2020, and that 12,590 women would die from EC, the estimated new and death cases increased 3,740 and 430 compared with that in 2019, relatively (1). Refer to the diagnosis of EC staging, although International Federation of Gynecology and Obstetrics (FIGO) remains the gold standard, about 33.33% of the tumor characterized with atypical morphology, resulting in the 10–37% inconsistency rate among different diagnoses, causing the current diagnostic criteria could not be uniformly applied practical work. The Cancer Genome Atlas (TCGA) classifies EC into four groups, each of which is based on different histopathology or molecular sub-type, as well as prognostic potential: group 1, polymerase epsilon (POLE), ultramutated, associated with good prognosis; group 2, microsatellite instability (MSI) hypermutated; group 3, copy-number low (CN-Low) endometrioid, group 2 and 3 shown similar progression-free survival rates; group 4, copy-number high (CN-High), serous-like, with worse prognosis. Also new biologic and molecular therapies for the treatment of endometrial carcinoma are being assessed in clinical trials. Application of TCGA classification may help in deciding the use of immunotherapy with immune checkpoint inhibitors like anti-programmed cell death-1 (PD-1)/programmed cell death-ligand 1 (PD-L1) treatment including lenvatinib/pembrolizumab for TMB-H [≥10 mutations/megabase (mut/Mb)] or MSI-high/mismatch repair (MMR) deficient tumors (the multicohort phase Ib KEYNOTE-028 study) (2-4). NCCN also recommended biomarker-directed systemic therapy for second-line treatment for EC like bevacizumab (randomized phase II trial Gynecologic Oncology Group trial) (5-7), nivolumab, larotrectinib or entrectinib for neurotrophin receptor kinase (NTRK) gene fusion-positive tumors (8). However, the reproducibility of the differences and associations between each molecular subtype and histological diagnosis is disputed among different researchers. Therefore, there is an increasing need to identify novel molecular biomarkers in order to achieve individualized treatment options. On the other side, for patients have no fertility requirements with high risk factors, surgery combined with radiotherapy, chemotherapy, systemic therapy, hormone therapy is still the main treatment at present (9). However, radio-chemoresistance remains a major obstacle in EC therapy (10), thus the development of a new anti-cancer therapeutic strategy and seeking effective biomarker is necessary.
In recent years, hydrogen therapy has been shown to be a promising therapeutic method for treating cervical cancer, breast cancer, cutaneous melanoma (11), ovarian cancer (12) and lung cancer (13). In our previous research (14), the consumption of hydrogen-enriched drinking water was shown to reduce endometrial tumor volume, density and weight in a xenograft mice model (Figure S1), suggesting that drinking hydrogen-rich water (HRW) might be served as an effective treatment or adjuvant therapy for EC, while the mechanism of how they function remain unknow. To characterize the effect of hydrogen water on signal transduction, substrate specificity, and peptidase functional efficiencies, a new approach called quantitative multiplex substrate profiling by mass spectrometry (qMSP-MS) (15) which utilizes isobaric tandem mass tags (TMTs) was utilized herein. TMTs are available with different reactive groups, which enables deep proteome coverage for multiple samples in a reasonable amount of time (16,17). Unlike studies that focus on RNA sequencing analysis, which can only detect gene expression alterations, or affymetrix single nucleotide polymorphisms (SNP) microarrays used in biospecimens to analyze mRNA, miRNA and methylation data which cost high, TMTs enables relative peptide and protein quantification across analyzed samples as means of identifying differential expression and can therefore provide insight into the proteolytic activities occurring within a complex biological sample (18,19).
When performing a MS-based analysis, differentially expressed proteins (DEPs) are identified and typically characterized using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, with differences further explored using cluster analysis (20), in which the sample and the variable is based on gathered quantitative data. GO analysis is used to describe the properties of gene products and is used to cluster DEPs based on the categories of biological process, molecular work and cellular component (21). KEGG pathway analysis is utilized to enrich pathways with a significant DEP involvement to better understand the biological processes in cells and mechanisms associated with traits, diseases, or drug response.
The main aim of this study was to perform a comprehensive survey of the proteome in a xenograft mouse model for EC to examine the molecular effects after HRW treatment. As such, mice were treated with either HRW or purified water [negative control (NC)], DEPs were identified using qMSP-MS, and the obtained data was examined using bioinformatics. All efforts were done to reveal new predictive DEPs in EC and can provide further insight into the molecular mechanisms of novel biomarkers to provide a more accurate framework of predicting the progress in EC, which would be useful to provide important information for prognosis and further response to additional adjuvant treatment options of EC. We present the following article in accordance with the REMARK reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2969).
Methods
Xenograft mouse model
Female BALB/c mice of 4 weeks of age, weighing 18–25 g, implanted with 1×107 luciferase-AN3CA cells on right shoulder were generated for xenograft mouse models of EC, and purchased from Shanghai Ling Chang BioTech Co., Ltd. affiliated to Shanghai Slake Laboratory Animal Co., Ltd. Experiments were performed under a project license (2020SQ301) granted by institutional board of the Ethical Committee on Human Research of Shanghai General Hospital affiliated to Shanghai Jiao Tong University, China, in compliance with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023) guidelines for the care and use of animals. BALB/c mice were housed under barrier environment, with each mouse in an independent ventilation cage, at 22–26 °C and humidity 45–65%, and were maintained on a 12/12-h light/dark cycle (lights on at 08:00 h). Mat and feed were changed every 2–3 days. The nine mice were randomly divided into two treatment groups: the HRW treatment group with the concentration of 1.0 ppm (n=6; H1–3) and the purified water (NC) group (n=3; P1–3) randomly when the tumors grew 3 to 4 mm in diameter and were visible. The mice were separately treated by gavage with either HRW or pure water control (20 mL/kg/d), and the mice were only allowed to drink the water associated with their group. We changed the HRW every 2 h each day, and the treatments were maintained for 24 days. We anesthetized and sacrificed the animals with an overdose of 2% sodium pentobarbital (0.5 mL), and then used cervical dislocation to confirm death, and took the subcutaneous tumorigenic tissue of mice from different groups. All samples were histologically diagnosed as EC by two independent pathologists. Both HRW intake was monitored throughout the experimental period and was nearly the same among groups.
Protein extraction
We chose six tumor samples for each group processed independently: three samples from HRW group labeled No. A1, A2, A3, three samples from NC group labeled No. B1, B2, B3. An equal amount of tumor sample was obtained, pooled and processed independently before labeling. The samples were then combined with double volume SDT buffer [4% sodium dodecyl sulfate (SDS), 100 mM Tris-HCl, pH 7.6], transferred to tubes containing lysing matrix A (MP Biomedicals, Solon, OH, USA) and homogenized using a FastPrep-24 5G system (MP Biomedicals; 24×2, 6.0 M/S, 60 s, two cycles). The samples were then sonicated (80 w, 10 s pulse, 10 s interval, 10 cycles) on ice and boiled for 10 min. The crude extract was centrifuged at 14,000 g for 15 min and the supernatants were collected. The supernatants were then filtered through Spin-X 0.22 µm filters (8160; Corning, Corning, NY, USA). Proteins were quantified using a bicinchoninic acid (BCA) Protein Assay kit (P0012; Beyotime, Shanghai, China) according to the manufacturer’s instructions and qualified via 12% SDS-polyacrylamide gel electrophoresis (PAGE) (P0015F, Beyotime; 250 V, 40 min), with 20 µg of protein combined with µg of 6x SDS sample buffer; this was then boiled for 5 min prior to loading. The obtained filtrates were then stored at –20 °C until further use.
Protein digestion
Protein digestion was performed using a filter-aided sample preparation (FASP) procedure (22). Briefly, 150 µg of total protein from each pool was diluted in 100 mmol dithiothreitol (DTT), boiled for 5 min, and cooled to 25 °C. To remove the low-molecular-weight components, samples were washed with 150 µL UA washing buffer (8 M Urea, 150 mM Tris-HCL, pH 8.5) and repeated ultrafiltration (Microcon units, 30 kD; Sartorius, Palaiseau, France, VN01H22), then performed at 12,500 g for 15 min. Next, samples were alkylated by adding 100 µL of indole-3-acetic acid (IAA) buffer (100 mM IAA in UA), followed by agitation (600 rpm) for 1 min and 30 min incubation in the dark at 25 °C. The samples were then centrifuged at 12,500 g for 15 min, the filtrate was discarded, and 100 µL of fresh UA washing buffer was added, followed by centrifugation. The filter was washed two times with 100 µL 40 nM NH4HCO3 (17837; Sigma-Aldrich, St. Louis, MO, USA) and centrifuged at 12,500 g for 15 min. Each sample was then digested in 4 µg of sequencing grade modified trypsin (V5117; Promega, USA) in 40 µL of 40 mM NH4HCO3, agitated (600 rpm) for 1 min, and incubated at 37 °C for 16–18 h. Next, the samples were centrifuged at 12,500 g for 15 min, the filter was washed with 20 µL of 40 nM NH4HCO3, and the samples were re-centrifuged. Samples were then incubated in 40 µL 0.1% tetraethylammonium bicarbonate (TEAB) buffer at 37 °C overnight. Finally, the resulting peptides were collected via centrifugation, and the concentrations were calculated at OD280.
TMT labeling
TMTs (Thermo Fisher Scientific) with different reporter ions (126–131 Da) were applied as isobaric tags for relative quantification. Samples were analyzed in each TMT 6-plex, an extra EBC sample was prepared as a reference standard. The pooled tumor sample was divided into eight equal samples. The standard sample in each TMT 6-plex was labeled as TMT-126, while other five test samples were labeled from TMT-127 to TMT-131. The balance sample was labeled as TMT-103, 102, 101, 100, 99, and 98, respectively. The peptide reaction sample connected the TMT reagent to the lysine and N-terminal amino acid residues on the peptide, and then six standard sample were connected to the peptide reaction sample through the balance sample, so that six kinds of relative molecular weights as 229 were formed. TMT labeling was performed according to the manufacturer’s instructions.
Briefly, transfer 100 µg per condition into a new tube and add 100 mM TEAB buffer to the protein solution to a final volume of 100 µL. Then 5 µL of 200 mM tris (2-carboxyethyl) phosphine (TCEP) was added into each sample and all samples were incubated at 55 °C for 1 hour. Next, each sample was added 5 µL of 375 mM iodoacetamide and incubated in the dark at room temperature for 30 mins. After precipitation and resuspension, proteins were digested overnight at 37 °C with 2.5 µg trypsin (Sigma, USA). The digested samples were individually labeled with TMT 6 reagents at room temperature for 1 h. At last, the labeled peptide aliquots were combined for subsequent fractionation. Labeled peptide were lyophilized, reconstituted, and fractionated with a C18 column (Waters, USA) by basic reversed-phase liquid chromatography (RPLC) method using a gradient of 5% to 95% solvent B (90% ACN, pH 10) in 40 min. A total of 40 fractions were collected and concatenated to 20 fractions, vacuum dried and stored at –80 °C until further analysis.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Each fraction was loaded onto a reverse phase trap column (Thermo Fisher Scientific; Acclaim PepMap 100, 50 µm × 15 cm, nanoViper, P/N164943) coupled with a C18-reversed phase analytical column (Thermo Scientific Easy Column, 10 cm long, 75 µm inner diameter, 3 µm resin) in buffer A (0.1% formic acid) and separated with a linear gradient of buffer B (84% acetonitrile and 0.1% formic acid) at a flow rate of 300 nL/min. The linear gradient proceeded as follows: 0–55% solution B for 80 min, 55–100% solution B for 5 min, and 100% solution B for 5 min.
Ten fractions from each sample were analyzed using a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled with a nanoflow high-performance liquid chromatography system (Easy nLC; Thermo Fisher Scientific) in positive ion mode for 90 min. The MS data was acquired using a data-dependent top 10 method that dynamically chooses the most abundant precursor ions from the survey scan (300–1,800 m/z) for higher-energy collisional dissociation (HCD) fragmentation. The instrument parameters were set as follows: automatic gain control target = 3e6, dynamic exclusion duration of 40 s, survey scan resolution of 700,000 (m/z 200), HCD spectra resolution of 35,000 (m/z 200) and an isolation width at m/z 2. The normalized collision energy was 30 eV and the underfill ratio was 0.1%. After performing LC-MS/MS, the data was evaluated using MASCOT 2.6 software (Matrix Science).
Protein identification and bioinformatics analysis
The acquired MS spectra were analyzed using the MASCOT search engine (version 2.6; Matrix Science, London, UK) within Proteome Discoverer 2.1 (Thermo Electron, San Jose, CA, USA). The parameters were set as follows: the enzyme used was trypsin, a peptide mass tolerance ±10 ppm, a fragment mass tolerance of 0.05 Da, and a max of 2 missed cleavages. Additionally, TMT sample was set as a fixed modification, and oxidation was set as a variable modification. The peptide false discovery rate (FDR) was set to ≤0.01 and all peptide ratios were normalized to the median protein ratio, with the median protein ratio defined as 1 after normalization.
The identified DEPs were blasted against the Mus Musculus (mouse) database using GO annotations were obtained by UniProt-GOA database (http://www.ebi.ac.uk/GOA/). Functional annotations were obtained using the KEGG database (http://geneontology.org/). GO and KEGG pathway enrichment were performed, with significance determined based on a two-tailed Fisher’s exact test (P<0.05). Hierarchical clustering analysis was performed using Cluster 3.0 (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm) and the Java Treeview (jtreeview.sourceforge.net). Protein-protein interaction (PPI) networks were constructed using Cytoscape and analyzed using the IntAct Molecular Interaction Database (http://www.ebi.ac.uk/intact/).
Hierarchical clustering
Further hierarchical clustering was performed based on the obtained functional classifications. First, all of the enriched categories, along with their P values, were sorted, and then filtered to isolate categories that were enriched in at least one of the clusters. The filtered P value matrix was then transformed by the function x = –log10, resulting in the x values being z-transformed for each functional category. The transformed data was then clustered using one-way hierarchical clustering in Genesis. Cluster membership was visualized using heatmaps and each protein category was examined in the InterPro database, with significance determined by using a two-tailed Fisher’s exact test (P<0.05).
Immunohistochemical (IHC) staining
IHC staining was conducted to exam kirsten rat sarcoma viral oncogenes homologue (KRAS), methionine sulfoxide reductase A (MSRA), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon (YWHAE) and trans-acting transcription factor 1 (SP1) expression in EC (n=17) and atypical hyperplasia (n=3) sections from hysterectomy, as well as secretory phase endometrium specimens (n=2) from curettage in Shanghai General Hospital affiliated to Shanghai Jiao Tong University, along with xenograft mice tumor tissue sections in the HRW and NC groups. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study methodologies were approved by the Ethical Committee on Human Research of Shanghai General Hospital affiliated to Shanghai Jiao Tong University (2020SQ301). All the participants have been informed consent before taking part. Xenograft tumor tissue was excised from mice sacrificed by cervical dislocation, endometrial specimens were randomly examined by two independent investigators. For IHC analysis, tissues were formalin-fixed and paraffin-embedded. Antigen was retrieved in citrate buffer (pH 6.0) after boiling, incubated with 0.01% Triton-X100 for 30 min, and 5% bovine serum albumin for 20 min. Anti-rabbit KRAS (1:50, Abcam, ab180772), anti-rabbit MSRA (1:25, Abcam, ab16803), anti-rabbit YWHAE (1:50, Abcam, ab43057) and anti-rabbit SP1 (1:50, Abcam, ab124804), primary antibodies were added overnight at 4 °C in a humidified chamber, co-incubated with biotinylated secondary antibody for 50-min. For quantitative analysis of immunohistochemistry, plaque images were visualized and analyzed with a microscopic imaging analysis system (IX71, Olympus Ltd., Japan). The IHC staining was scored for both positive cells’ proportion (0 score: 0%, 1 score: ≤10%, 2 score: 11–50%, 3 score: 51–80%, and 4 score: ≥80%) and staining intensity (0 score: negative, 1 score: weak, 2 score: moderate, and 3 score: strong), which ultimately resulted in designations of complete loss of expression, or weak, moderate, or strong expression, respectively. KRAS, MSRA and YWHAE was mainly stained in cytoplasmic staining, SP1 was mainly stained in Nuclear.
Statistical analysis
For the identification of DEPs, a fold-change (FC) cutoff of >1.2 or <0.83 was established and significance was determined using a Student’s t-test. When performing GO and KEGG pathway analyses, significance was determined using a two-tailed Fisher’s Exact Test. For all statistical evaluations, significance was determined as P <0.05.
Results
HRW treatment inhibits endometrial tumorigenesis in vivo
We had verified that drinking HRW inhibited tumor growth in a BALB/c mice model of EC which were implanted with 1×107 luciferase-AN3CA cells on the right shoulder of the animal in our previous research (14) (Figure S1A). All six BALB/c mice were fed with either HRW with the concentration of 1.0 ppm (HRW: H1–6) or purified normal control (NC: P1–3) water ad lib each day. In order to balance individual differences, tumor volume on the first day was defined as 1 and the durations of increased or decreased proportions were recorded as the relative tumor volume. We observed a decreased trend in the relative tumor volume (mm3) and mice weight (g) in the HRW group on all or most the observed days, compared with the control group (Figure S1B,C). There was also a decrease trend in total radiance (ROI) (p/sec/cm2/sr) which indicated lower tumor density in the HRW group on day 12, 13, 24, compared with the control group (P<0.05) (Figure S1D,E).
Mass spectrometry
We next chosen the tumor sample A1–3 (HRW group) and B1–3 (NC group) for TMTs exam, respectively. Prior to examination, the obtained protein samples were quantified (BSA assay) and qualified (SDS-PAGE) and showed good protein quality, sufficient quantity and consistency between samples. A recovery rate of peptides/proteins to the total initial quantity of protein is 45%. The spectra data was examined against the UniProt Mus Musculus database to identify protein groups (unique peptide >2), proteins and peptides (Table 1 and Figure S2). Furthermore, comparisons between the peptide theoretical molecular weight and the experimentally determined molecular weight were also examined in MASCOT and results were displayed a normal distribution (Figure S2A). Additionally, ions score distributions were evaluated with a higher peptide score associated with a higher match to the theoretical peptide (Figure S2B). The molecular weight distribution had a range between 0 and over 250 kDa (Figure S2C) and the isoelectric point distribution had a range between 1 and 14 (Figure S2D). The peptide length distribution was also examined and reflected cleavage at lysine and arginine (Figure S2E). The protein sequence coverage distribution showed the percentage of protein sequences that align to the identified peptides (Figure S2F). The peptide count distribution reflected the number of distinct sequences within a protein group (Figure S2G). The protein ratio distribution evaluated the log2 (FC) of the experimental group when compared to the control protein group, and displayed a normal distribution (Figure S2H).
Table 1
| Sample group | HRW | Pure water | |||||
|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | ||
| Quantitative result | |||||||
| Concentration (μg/μL) | 13.9 | 17.2 | 15.6 | 12.1 | 13.0 | 15.7 | |
| Volume (μL) | 300 | 300 | 300 | 300 | 300 | 300 | |
| Total amount (μg) | 4,170 | 5,160 | 4,680 | 3,630 | 3,900 | 4,710 | |
| Sample evaluation | a | a | a | a | a | a | |
| Mass spectrometry result | |||||||
| Database | Uniprot_MusMusculus_16998_20180905 | ||||||
| Protein group | 2,015 | 1,983 | 1,957 | 2,029 | 2,022 | 1,968 | |
| Protein group (unique peptide >2) | 1,233 | 1,244 | 1,260 | 1,263 | 1,243 | 1,254 | |
| Mass spectrometry evaluation | A | A | A | A | A | A | |
Quantitative and SDS-PAGE results showed that the protein was of good quality, sufficient total amount, and have good parallelism between samples. Protein pre-mass spectrometry showed normal digestion and chromatographic mass spectrometry. HRW, hydrogen-rich water.
Enrichment-based clustering protein domain significant difference analysis
The aim of this project was to use an integrated approach combining TMT labeling and LC-MS/MS to quantify dynamic changes within the proteome of endometrial tumors exposed to HRW treatment. After performing a quality validation, LC-MS/MS was performed and a total of 427,451 (97,489 matched) spectra were obtained. Of these spectra, a total of 51,071 peptides, including 47,027 unique peptides and 6,974 proteins, were detected across all the samples with an average peptide mass error <10 ppm. Average A/B (mean value of group A/B) displayed difference comparison group, proteins that meet the screening fold greater than 1.2-fold (up-regulation) or less than 0.83-fold (down-regulation), which means P value (t-test) less than 0.05 are considered differential expression proteins. Based on the criterion to a total of 57 DEPs, 11 significantly DEPs were identified in the HRW group relative to the NC group. Among the identified DEPs, 5 were up-regulated and average A/B were shown, including gata zinc finger domain containing 1 (Gatad1) (FC =1.497), tweety homologue (Ttyh3) (FC=1.411), nima‐related kinase‐4 (Nek4) (FC =1.396), dual-specificity tyrosine-regulated kinase 2 (Dyrk2) (FC =1.293) and GTPases of the immunity associated proteins 1 (Gimap1) (FC =1.222), while 6 were down-regulated, including SP1 (FC =0.811), male specific lethal 1 (Msl1) (FC =0.796), pleckstrin homology domain containing, family A member 7 (Plekha7) (FC =0.752), DTW domain containing 2 (Dtwd2) (FC =0.748), KRAS (FC =0.649), and MSRA (FC =0.547). The K-means clustering for these DEPs was visualized using a heatmap (Figure 1A, Table 2, Table S1).
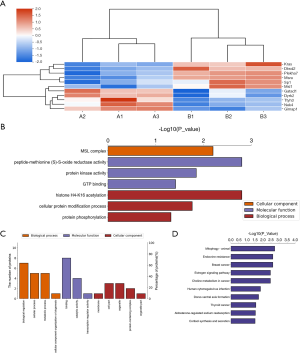
Table 2
| Function | Accession | Gene name | Average A | Average B | Average A/B | P value |
|---|---|---|---|---|---|---|
| Up-regulated | Q920S3 | Gatad1 | 119.9 | 80.1 | 1.4968789 | 0.03311027 |
| Q6P5F7 | Ttyh3 | 117.033333 | 82.9666667 | 1.41060667 | 0.04967401 | |
| Q9Z1J2 | Nek4 | 116.533333 | 83.4666667 | 1.39616613 | 0.01765495 | |
| Q5U4C9 | Dyrk2 | 112.766667 | 87.2333333 | 1.29270157 | 0.02412443 | |
| P70224 | Gimap1 | 110 | 90 | 1.22222222 | 0.0038861 | |
| Down-regulated | O89090 | SP1 | 89.6 | 110.433333 | 0.81134923 | 0.04827976 |
| Q6PDM1 | Msl1 | 88.6333333 | 111.366667 | 0.7958695 | 0.02366069 | |
| Q3UIL6 | Plekha7 | 85.8666667 | 114.133333 | 0.75233645 | 0.00498638 | |
| Q9D0U1 | Dtwd2 | 85.5666667 | 114.466667 | 0.74752475 | 0.00630704 | |
| P32883 | KRAS | 78.7 | 121.3 | 0.64880462 | 0.00986105 | |
| Q9D6Y7 | MSRA | 70.7 | 129.3 | 0.54679041 | 0.00242538 | |
| – | P62259 | YWHAE | 100.1 | 99.86667 | 1.002336 | 0.977884 |
Average A/B, the mean value of group A/B, means difference comparison group, proteins that meet the screening fold greater than 1.2-fold (up-regulation) or less than 0.83-fold (down-regulation) and P value (t-test) less than 0.05 are considered differential expression proteins. DEP, differentially expressed protein; HRW, hydrogen-rich water.
Bioinformatics analysis on GO annotation
To characterize the functions of the significant DEPs associated with hydrogen treatment, GO annotations were enriched. This classification system provides dynamically updated descriptions of gene and gene product properties from three perspectives: the biological process, molecular function and cellular component (23). GO annotations were derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). For each category, enriched DEPs we compared to all the identified proteins using a two-tailed Fisher’s exact test, focusing on the top 12 rankings for each category, Figure 1B showed Enrichment analysis of GO term performed with 11 differential proteins displayed by –log10 (P value) (Tables S2,S3) (P<0.05). In the biological process category, 63.64% of proteins were associated with biological regulation (SP1, Msl1, Nek4, Gatad1, KRAS, MSRA and Dyrk2), 45.45% with cellular and metabolic processes (Msl1, Nek4, KRAS, MSRA and Dyrk2), and 9.09% with cellular component organization or biogenesis (Msl1). In the molecular function category, 72.73% of proteins were associated with binding activity (SP1, Plekha7, Nek4, Gatad1, MSRA, KRAS, Gimap1, and Dyrk2), 36.36% with catalytic activity (MSRA, KRAS, Nek4, and Dyrk2), and 9.09% with transcription regulator activity (SP1). In the cellular component category, 27.27% of proteins were involved in cell parts and organelles (SP1, Msl1 and KRAS), 18.18% were involved in the protein-containing complex (SP1, Msl1), and 9.09% were involved in membrane (KRAS) and organelle parts (Msl1) (Figure 1C, Table 3). Overall, the categories of binding region and biological regulation ranked the highest in association with hydrogen treatment in EC.
Table 3
| GO type | GO name | Sequence name | Percentage of proteins (%) | Term | TestSeqs | –Log10(P value) |
|---|---|---|---|---|---|---|
| Biological process | Biological regulation | SP1, Msl1, Nek4, Gatad1, KRAS, MSRA, Dyrk2 | 63.64 | Histone H4-K16 acetylation | Msl1 | 2.80209 |
| Cellular process | Msl1, Nek4, KRAS, MSRA, Dyrk2 | 45.45 | Cellular protein modification process | MSRA | 1.76381 | |
| Metabolic process | Msl1, Nek4, KRAS, MSRA, Dyrk2 | 45.45 | Protein phosphorylation | Nek4, Dyrk2 | 1.3205 | |
| Cellular component organization or biogenesis | Msl1 | 9.09 | Regulation of transcription by RNA polymerase II | SP1 | 1.21215 | |
| – | – | – | Regulation of transcription, DNA-templated | SP1 | 0.78717 | |
| – | – | – | Signal transduction | KRAS | 0.63045 | |
| Molecular function | Binding | SP1, Plekha7, Nek4, Gatad1, MSRA, KRAS, Gimap1, Dyrk2 | 72.73 | Peptide-methionine (S)-S-oxide reductase activity | MSRA | 2.80209 |
| Catalytic activity | MSRA, KRAS, Nek4, Dyrk2 | 36.36 | Protein kinase activity | Nek4, Dyrk2 | 1.84984 | |
| Transcription regulator activity | SP1 | 9.09 | GTP binding | KRAS | 1.4167 | |
| – | – | – | Sequence-specific DNA binding | Gatad1 | 1.06359 | |
| – | – | – | DNA binding transcription factor activity | SP1 | 0.85774 | |
| – | – | – | GTPase activity | KRAS | 0.6617 | |
| – | – | – | ATP binding | Nek4, Dyrk2 | 0.62254 | |
| – | – | – | Zinc ion binding | Gatad1 | 0.54221 | |
| – | – | – | Nucleic acid binding | SP1 | 0.42896 | |
| – | – | – | Protein binding | Plekha7 | 0 | |
| Cellular component | Membrane | KRAS | 9.09 | Membrane | KRAS | 0.65919 |
| Cell part | SP1, Msl1, KRAS | 27.27 | – | – | – | |
| Organelle | SP1, Msl1, KRAS | 27.27 | Nucleus | SP1 | 0.44002 | |
| Protein-containing complex | SP1, Msl1 | 18.18 | Transcription factor complex | SP1 | 0.69604 | |
| Organelle part | Msl1 | 9.09 | MSL complex | Msl1 | 2.20096 |
GO, Gene Ontology.
KEGG pathway annotation
KEGG pathway annotations were performed using the identified DEPs, with significance determined based on a two-tailed Fisher’s exact test (P<0.05). The results identified a total of 87 pathways. The top 10 pathways were identified ranged by the –log10 (P value) (Figure 1D, Table 4, Table S4) and included mitophagy animal, endocrine resistance, breast cancer, estrogen signaling pathway and choline metabolism in cancer, human cytomegalovirus infection, dorso-ventral axis formation, thyroid cancer, aldosterone-regulated sodium reabsorption and cortisol synthesis. The DEPs associated with these pathways included SP1 and KRAS, indicating that they may be key proteins that are able to disrupt multiple pathways.
Table 4
| Map name | DiffSeqs | –Log(P value) |
|---|---|---|
| Mitophagy—animal | SP1, KRAS | 2.72546 |
| Endocrine resistance | SP1, KRAS | 2.6104 |
| Breast cancer | SP1, KRAS | 2.6104 |
| Estrogen signaling pathway | SP1, KRAS | 2.46304 |
| Choline metabolism in cancer | SP1, KRAS | 2.46304 |
| Human cytomegalovirus infection | SP1, KRAS | 1.84285 |
| Dorso-ventral axis formation | KRAS | 1.72633 |
| Thyroid cancer | KRAS | 1.55211 |
| Aldosterone-regulated sodium reabsorption | KRAS | 1.52894 |
| Cortisol synthesis and secretion | SP1 | 1.50697 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Analysis of protein interaction networks
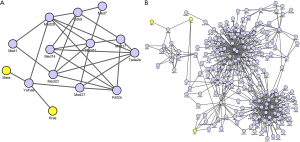
To further characterize the relationship between identified DEPs, PPI networks were constructed using Cytoscape Software (version 3.6.1) (Figure 2). Both the identified DEPs in mice and corresponded DEPs in human were displayed. Adjacent proteins were assumed to have similar clustering, which was composed of individual proteins through their interactions to participate in all aspects of life processes such as biological transmission, gene expression regulation, energy and material metabolism, and cell cycle regulation. In the PPI network graph, KRAS and MSRA, both down-regulated, were found to interact with YWHAE directly (FC =1.0023, P=0.9778; Figure 2A). Once this interaction was expanded to a larger network in human, SP1 (down-regulated) was also incorporated (Figure 2B). However, KRAS-YWHAE-MSRA was more closely connected in mice than in human PPI network, which need further explore.
TCGA database comparison and survival analysis of DEPs
In order to vertify the comparison and survival analysis the above DEPs, the identified DEPs (n=11) and YWHAE were examined against TCGA database analyzed by the online software UALCAN using Kaplan-Meier Plotter (http://ualcan.path.uab.edu/analysis.html), with protein expression between EC and normal endometrial tissue examined, including correlations with stage, menopause status, histology and age (24,25). When examining the significantly down-regulated DEGs, the TCGA database showed a significant difference between endometrial tumors and normal endometrium for KRAS (P=6.06E–03), YWHAE (P=2.22E–16), SP1 (P=4.17E–09) and MSRA (P=4.47E–05) expression. The 5-year survival rate of EC patients between low or moderate versus high expression levels was also examined: KRAS (0.8 vs. 0.75, P=0.64), YWHAE (0.8 vs. 0.75, P=0.59), SP1 (0.75 vs. 0.73, P=0.95) and MSRA (0.75 vs. 0.7, P=0.91) (Figure S3). Thus, KRAS, YWHAE, SP1 and MSRA were chosen to conduct IHC assay to validate their expression in EC tissue.
Clinical pathological factors associated with KRAS, SP1, YWHAE and MSRA
We stained human endometrial tissue sections selected among the patients in our hospital: EC (n=17), atypical hyperplasia (n=3) and secretory phase endometrium (n=2), and mice tumor tissue sections in the HRW (n=3) and NC (n=3) groups for KRAS, YWHAE, SP1 and MSRA expression. IHC expression pattern of KRAS, SP1, YWHAE and MSRA protein in all tissue sections of EC and histopathological clinical evaluations were presented in Figure 3. Strong positive staining of KRAS, YWHAE, SP1 and weak staining of MSRA were found in atypical hyperplasia endometrial tissue (3/3). The percentage of moderate to strong positive expression of KRAS, YWHAE and SP1 in EC specimens were 82.35% (14/17), 88.23% (15/17) and 82.35% (14/17), while weak expression of MSRA accounted for 88.23% (15/17) in EC specimens, respectively. Collectively, weak or negative expression of the four proteins were shown in secretory phase endometrium specimens (n=2) (Figure 3A). Most of the EC patients were observed in the age group of 44–83 years. The clinical evaluations of EC patients presented that out of 17 cases, 16 cases were identified pure endometrioid carcinoma, another case was serous carcinoma. No of the patient had parametrial, adnexa, endocervical glandular, cervical stromal connective tissue, and lymph node invasion. High risk factors include age over 50 years old, histologic grade 2 or 3, deeper myoinvasion (>50%), lymph-vascular space invasion (LVSI). The frequency of myometrial invasion was high (15/17, 88.24%), the mean depth of myometrial invasion was 0.45±0.21 cm (Figure 3B). The histopathological type of EC patients: grade 1: nine patients, grade 2: three patients; grade 3: five patients. Compared between the moderate to strong and negative to weak expression cases of KRAS, YWHAE, SP1 and MSRA, there was significantly difference between the expression group of CK7, CD10.
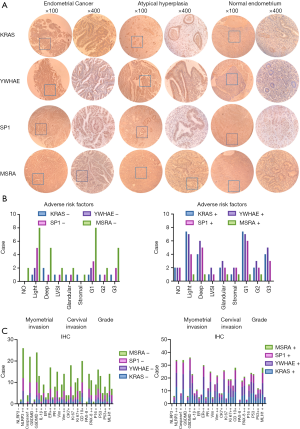
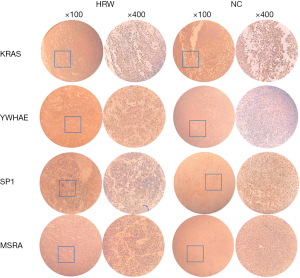
Paired box-8 (PAX-8), Mutl homolog l gene (MLH1) (KRAS: P=0.0001, YWHAE: P=0.0006, SP1: P=0.0273, MSRA: P=0.0003), and the expression group of P16 and P53 (KRAS: P=0.0157, YWHAE: P=0.0104, SP1: P=0.0041, MSRA: P=0.0104), as well as the expression group of Vim and Ki67 (YWHAE: P=0.0036, MSRA: P=0.0116) (Figure 3C, Table S5). Also, the expression of KRAS, YWHAE and SP1 protein were moderate to strong positive, and the expression of MSRA were weak or negative in HRW group in xenograft tumor tissue compared with that in NC group (Figure 4). However, since the samples size was limited, further samples should be collected to draw statistical conclusions.
Discussion
TMT labeling coupled with LC-MS/MS offers a robust approach that allows relative protein abundances for thousands of proteins in multiple samples to be identified simultaneously due to each isobaric compound containing a different number of heavy isotopes in their mass-reporter region. Furthermore, TMT labeling can greatly improve reproducibility while providing protein identification and relative quantification simultaneously (22). Herein, TMTs were used to quantify cleaved and uncleaved peptides within endometrial tumors harvested from mice treated with HRW, in order to be compared to profiles from the control samples. The results identified five up-regulated (Gatad1, Ttyh3, Nek4, Dyrk2, and Gimap1) and six down-regulated (Plekha7, Dtwd2, KRAS, Msl1, SP1, and MSRA) DEPs. These identified DEPs were then further analyzed using GO, KEGG, MASCOT, TCGA databases and Cytoscape as well as IHC.
Among the down-regulated proteins, KRAS is a member of the RAS superfamily proteins and a proto-oncogene (gene ID: 3845) located at chromosome 12 (12p 12.1) which encodes 21 kDa protein, it is primarily involved in the cellular response to extracellular signals. KRAS mutants in codon 12 frequently with an alteration of guanine to adenine (G>A) (26). KRAS mutations promote the down-regulation of membrane receptor signaling through mitogen-activated protein kinase (MAPK) and phosphoinositide-3-kinase/v-akt murine thymoma viral oncogene (PI3K/AKT) pathways, which result in dependent autophagy that is necessary for cancer progression as well as promote proliferation and subsequent carcinogenesis (27-32). KRAS-driven tumor regression and growth impaired was observed after nanoparticle-mediated delivery of siKRAS to KRAS-Mutant tumors in a mouse model (33,34), KRAS mutants knock-out leads to inhibition of upstream signaling pathways (35,36). Further to this, KRAS mutations appear to be a stage ahead of TP53 involvement and clonal expansion (37,38). There is also a positive relationship between the KRAS gene and estrogen receptors (ER) (39), MSI-positive and a molecular assessment of the depth of myometrial invasion of EC, which are generally thought to occur early in the EC pathway (40,41). KRAS mutations are present in 6–16% of endometrial hyperplasia specimens, 88% complex atypical hyperplasia and 10–30% type I estrogen-related EC (37,40,42). An increase in KRAS expression has been associated with a poor outcome in 3% of the primary and 18% of metastatic lesions, and an aggressive phenotype (43). There is a clear trend in the literature showing that KRAS plays the important role of predicting early checkpoint of transition from hyperplastic endometrium to early-stage well-differentiated (grade I) estrogen-related EC, as well as further transition from low- to high-grade type I EC (44-46).
The research progress of EC-related tumor biomarkers may carry important advantages in clinical practice of possible targeted therapy to EC patients in order to improve patient prognosis. A series of studies were devoted to inhibit KRAS mutations including KRAS direct binding molecules, KRAS membrane localization targeting enzymes, or downstream signaling (47,48), synthetic lethal interactors, inhibiting KRAS gene expression, though immune system pathways (49,50). Recent studies have also shown that a combination therapy of mitogen-activated extracellular kinase (MEK) inhibitors plus anti-estrogen agents may alter estrogen signaling in KRAS-mutant EC and thus improve the response rate (51). Herein, KRAS expression was significantly decreased in response to HRW treatment in xenograft mouse model (FC =0.649, P=0.009). Furthermore, GO annotation was associated with KRAS molecular and cellular functions, while KEGG analysis associated it with several cancer-related metabolic and hormonal pathways. Additionally, the IHC staining of KRAS in atypical hyperplasia endometrial tissue was stronger than that in EC specimens. When examining KRAS in the TCGA database, differential expression between EC and normal endometrial tissues was noted (P=6.06E–03), with the survival rate between low/moderate and high expression almost reaching a significant level (0.8 vs. 0.75, P=0.64). Based on this, KRAS status could be regarded as a potential prognostic marker, both in terms of transition from pre-malignant to malignant cell status, as well as progression from early to more advanced invasive cancer (52). The SP1 transcription factor contains a zinc finger motif and binds to glypican (GpC)‐rich promoter regions (53), it has been shown to act as either a promoter or repressor during cellular progression and proliferation (54,55). Dysregulation of SP1 has been found to be involved in many cancers, and results in the suppression of cell migration and invasion in squamous cervical cancer (56), promotes the migration of ovarian cancer cells (57) as well as stimulated stem cell of colon cancer growth and induce apoptosis (58). Additionally, SP1 knockdown could reverse the effects of miR-490 inhibition on the malignant behaviors of ishikawa cells and inhibited PI3K/AKT pathway which elucidated the roles SP1 axis in EC development and may provide a new strategy for EC therapy (59). In our study, SP1 expression decreased in response to HRW treatment (FC =0.811, P=0.048). Furthermore, the GO annotation associated SP1 with transcription and other cellular functions, while KEGG analysis associated it with cancer, metabolism, and estrogen associated pathways. When examining SP1 in the TCGA database and conducted by IHC, SP1 was differentially expressed when comparing EC and normal endometrial tissues (P=4.17E–09).
MSRA is located on chromosome 8p23.1 and encodes the methionine sulfoxide reductase, which is known to protect proteins from oxidation and acts as a reactive oxygen species (ROS) scavenger (60). MSRA performed a tumor-suppressive effect in both lung squamous carcinoma and adenocarcinoma than in adjacent normal tissues (61). However, there is no literature report on the expression of MSRA in EC in recent years. We demonstrated that MSRA were down-regulated DEPs detected in our study and displayed weak expressed in EC specimens by IHC.
To examine how these proteins potentially interact, Cytoscape was utilized to construct interaction networks which was through the interaction of each other protein to form a macromolecular complex to complete its biological functions, such as genetic material replication, gene expression regulation, cell signal transduction, metabolism, cell proliferation, apoptosis and so on. Both KRAS and SP1 are associated with several cell membrane receptors and act as signal transduction molecules. Additionally, we showed that KRAS was interacted with MSRA though YWHAE directly.
YWHAE (14-3-3ε) belongs to the 14-3-3 protein family, which are highly conserved from yeast to human and consist of seven mammalian isoforms (β, γ, ζ, η, θ, σ, and ε) with unique expression patterns in different cell types (62). YWHAE functions as a molecular framework to coordinate cellular signaling by binding to phosphoserine- or phosphothreonine-containing proteins (63,64). As to uterus tumor, YWHAE-NUTM2A/B endometrial stromal sarcomas (ESS) is a recently described variant of high-grade endometrial stromal sarcomas (HG-ESS) which is included in the 2014 WHO Classification of Tumors of the Female Reproductive Organs excluded from the prior WHO 2003 Classification (63,64). YWHAE interactome in myeloma cells also revealed enrichment in PI3K-AKT-mTOR. YWHAE were moderate to strong positive expressed in EC specimens by IHC in our research and TCGA database.
14-3-3 proteins interact with many binding partners, including αSyn, affecting cell cycle and transcriptional control, signal transduction, intracellular trafficking, and regulation of ion channels (65). YWHAE lncRNA down- or up-regulation induced corresponding a significant down- or up-regulation of KRAS gene at the mRNA level, YWHAE encoded lncRNA promotes activation of KRAS/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells (66). Starbase data also showed positive correlation between YWHAE gene and KRAS gene in colorectal cancer tissues. Specifically, positive effect of MSRA-dependent interaction on the ubiquitination of 14-3-3ζ through MSRA knockout mice exhibiting high levels of 14-3-3ζ compared with the corresponding wild-type strain (67,68). KRAS, SP1 and YWHAE are all associated with PI3K/AKT/mTOR pathway which are found implicated in EC pathogenesis, we hypothesized MSRA stimulated YWHAE, and YWAHE activated KRAS (Figure 5). On the other hand, both KRAS and YWHAE can be used as markers to evaluate tumor prognosis. KRAS mutations can cause resistance to epidermal growth factor receptor (EGFR) inhibitors (69). Thus KRAS mutation has emerged as the major negative predictive biomarker for response to anti-EGFR chemotherapy agents in colorectal cancer patients (29,70). YWHAE-NUTM2A/B fusion subsequent to a t(10;17) (q22;p13) has been associated with a more aggressive neoplasm and a poorer prognosis when compared to its low-grade counterpart in HG-ESSs (71). YWHAE translocation correlated with low mitotic index and improved prognosis of undifferentiated uterine sarcomas (72). YWHAE (14-3-3ε) expression is predictor of clinical outcome in a large dataset of myeloma patients receiving bortezomib (BTZ) as first line therapy (62). The 5-year survival rate of EC patients between low or moderate versus high expression levels was also examined by TCGA: KRAS (0.8 vs. 0.75, P=0.64), YWHAE (0.8 vs. 0.75, P=0.59), SP1 (0.75 vs. 0.73, P=0.95), which predicted that the lower risk in prognosis of EC in the low-expression group than in the high-expression group.
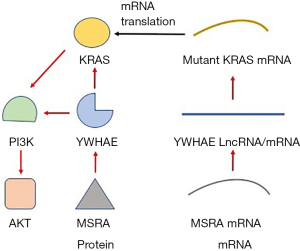
Conclusions
In conclusion, based on the previous discussion, it would be interesting to conduct a prospective study to delineate the role of the DEPs like KRAS, MSRA, SP1 and YWHAE as focused biomarkers, and even regarding KRAS in predicting standard individual treatment approach of cancer progression after hyperplasia with or without atypia in endometrium. Clinicopathological IHC staining could be conducted to exam these biomarkers in patients’ endometrial tissue to develop predictive outcome. However, how KRAS and MSRA interact with YWHAE, and the exact mechanism of how they affect EC requires further exploration. Additionally, one limiting factor of this study is the small sample size for the EC xenograft mouse model. Nevertheless, the biomarkers might provide a biased view as it may be optimistic to explain a complex carcinogenesis progression using the genes. Thus, future research should aim to validate KRAS, MSRA, SP1 and YWHAE as DEPs in subcutaneous tumorigenic tissue and determine their role in cell death in EC.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2969
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tcr-20-2969
Peer Review File: Available at http://dx.doi.org/10.21037/tcr-20-2969
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2969). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study methodologies were approved by the Ethical Committee on Human Research of Shanghai General Hospital affiliated to Shanghai Jiao Tong University (2020SQ301). All the participants have given the informed consent before taking part. Experiments were performed under a project license (2020SQ301) granted by institutional board of the Ethical Committee on Human Research of Shanghai General Hospital affiliated to Shanghai Jiao Tong University, China, in compliance with the National Institutes of Health guide for the care and use of Laboratory Animals (NIH Publications No. 8023) guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. [Crossref] [PubMed]
- Piulats JM, Guerra E, Gil-Martín M, et al. Molecular approaches for classifying endometrial carcinoma. Gynecol Oncol 2017;145:200-7. [Crossref] [PubMed]
- Kandoth C, Schultz N, Cherniack AD, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Simpkins F, Drake R, Escobar PF, et al. A phase II trial of paclitaxel, carboplatin, and bevacizumab in advanced and recurrent endometrial carcinoma (EMCA) - ScienceDirect. Gynecol Oncol 2015;136:240-5. [Crossref] [PubMed]
- Rose PG, Ali S, Moslemi-Kebria M, et al. Paclitaxel, carboplatin, and bevacizumab in advanced and recurrent endometrial carcinoma. Int J Gynecol Cancer 2017;27:452-8. [Crossref] [PubMed]
- Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409-13. [Crossref] [PubMed]
- Clinical Practice Guidelines in Oncology Uterine Neoplasms version 1. 2021 Available online: www.NCCN.org
- Onodera Y, Nam JM, Mei H, et al. Arf6-driven cell invasion is intrinsically linked to TRAK1-mediated mitochondrial anterograde trafficking to avoid oxidative catastrophe. Nat Commun 2018;9:2682-97. [Crossref] [PubMed]
- Montero J, Sarosiek KA, DeAngelo JD, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 2015;160:977-89. [Crossref] [PubMed]
- Zhao P, Jin Z, Chen Q, et al. Local generation of hydrogen for enhanced photothermal therapy. Nat Commun 2018;9:4241-52. [Crossref] [PubMed]
- Shang L, Xie F, Li J, et al. Therapeutic potential of molecular hydrogen in ovarian cancer. Transl Cancer Res 2018;7:988-95. [Crossref]
- Wang D, Wang L, Zhang Y, et al. Hydrogen gas inhibits lung cancer progression through targeting SMC3. Biomed Pharmacother 2018;104:788-97. [Crossref] [PubMed]
- Yang Y, Liu PY, Bao W, et al. Hydrogen inhibits endometrial cancer growth via a ROS/NLRP3/caspase-1/GSDMD-mediated pyroptotic pathway. BMC Cancer 2020;20:28-46. [Crossref] [PubMed]
- Guan L, Zhao M, Qian Y, et al. Phenotypic analysis combined with tandem mass tags (TMT) labeling reveal the heterogeneity of strawberry stolon buds. BMC Plant Biol 2019;19:505-29. [Crossref] [PubMed]
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature 2003;422:198-207. [Crossref] [PubMed]
- Leichert LI, Gehrke F, Gudiseva HV, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci U S A 2008;105:8197-202. [Crossref] [PubMed]
- Paek J, Kalocsay M, Staus DP, et al. Multidimensional Tracking of GPCR Signaling via Peroxidase-Catalyzed Proximity Labeling. Cell 2017;169:338-49.e11. [Crossref] [PubMed]
- Mertins P, Mani DR, Ruggles KV, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016;534:55-62. [Crossref] [PubMed]
- Götz S, García-Gómez JM, Terol J, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 2008;36:3420-35. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Wiśniewski JR, Zougman A, Nagaraj N, et al. Universal sample preparation method for proteome analysis. Nat Methods 2009;6:359-62. [Crossref] [PubMed]
- Gene Ontology Consortium. going forward. Nucleic Acids Res 2015;43:D1049-56. [Crossref] [PubMed]
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46:D956-63. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Sah B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Rosty C, Young JP, Walsh MD, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol 2013;26:825-34. [Crossref] [PubMed]
- Miller KA, Yeager N, Baker K, et al. Oncogenic Kras requires simultaneous PI3K signaling to induce ERK activation and transform thyroid epithelial cells in vivo. Cancer Res 2009;69:3689-94. [Crossref] [PubMed]
- Polosukhina D, Love HD, Correa H, et al. Functional KRAS mutations and a potential role for PI3K/AKT activation in Wilms tumors. Mol Oncol 2017;11:405-21. [Crossref] [PubMed]
- Duffy MJ, Lamerz R, Haglund C, et al. Tumor markers in colorectal cancer, gastric cancer and gastrointestinal stromal cancers: European group on tumor markers 2014 guidelines update. Int J Cancer 2014;134:2513-22. [Crossref] [PubMed]
- Todoric J, Antonucci L, Di Caro G, et al. Stress-activated NRF2-MDM2 cascade controls neoplastic progression in pancreas. Cancer Cell 2017;32:824-39.e8. [Crossref] [PubMed]
- Perera RM, Svetlana S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature 2015;524:361-5. [Crossref] [PubMed]
- Guo JY, Karsli-Uzunbas G, Mathew R, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447-61. [Crossref] [PubMed]
- Yuan TL, Fellmann C, Lee CS, et al. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov 2014;4:1182-97. [Crossref] [PubMed]
- Xue W, Dahlman JE, Tammela T, et al. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A 2014;111:E3553-61. [Crossref] [PubMed]
- Holderfield M, Deuker MM, Mccormick F, et al. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer 2014;14:455-67. [Crossref] [PubMed]
- Young A, Lou D, Mccormick F. Oncogenic and wild-type ras play divergent roles in the regulation of mitogen-activated protein kinase signaling. Cancer Discov 2013;3:112-23. [Crossref] [PubMed]
- Llobet D, Pallares J, Yeramian A, et al. Molecular pathology of endometrial carcinoma: practical aspects from the diagnostic and therapeutic viewpoints. J Clin Pathol 2009;62:777-85. [Crossref] [PubMed]
- Duggan BD, Felix JC, Muderspach LI, et al. Early mutational activation of the c-Ki-ras oncogene in endometrial cancer. Cancer Res 1994;54:1604-7. [PubMed]
- Tu Z, Gui L, Wang J, et al. Tumorigenesis of K-ras mutation in human endometrial carcinoma via upregulation of estrogen receptor. Gynecol Oncol 2006;101:274-9. [Crossref] [PubMed]
- Zauber P, Denehy TR, Taylor RR, et al. Strong correlation between molecular changes in endometrial carcinomas and concomitant hyperplasia. Int J Gynecol Cancer 2015;25:863-8. [Crossref] [PubMed]
- Alexander-Sefre F, Salvesen HB, Ryan A, et al. Molecular assessment of depth of myometrial invasion in stage I endometrial cancer: a model based on K-ras mutation analysis. Gynecol Oncol 2003;91:218-25. [Crossref] [PubMed]
- Alomari A, Abi-Raad R, Buza N, et al. Frequent KRAS mutation in complex mucinous epithelial lesions of the endometrium. Mod Pathol 2014;27:675-80. [Crossref] [PubMed]
- Salvesen HB, Carter SL, Mannelqvist M, et al. Integrated genomic profiling of endometrial carcinoma associates aggressive tumors with indicators of PI3 kinase activation. Proc Natl Acad Sci U S A 2009;106:4834-9. [Crossref] [PubMed]
- Tsuda H, Jiko K, Yajima M, et al. Frequent Occurrence of c-Ki-ras Gene Mutations in Well Differentiated Endometrial Adenocarcinoma Showing Infiltrative Local Growth with Fibrosing Stromal Response. Int J Gynecol Pathol 1995;14:255-9. [Crossref] [PubMed]
- Dobrzycka B, Terlikowski SJ, Mazurek A, et al. Mutations of the KRAS oncogene in endometrial hyperplasia and carcinoma. Folia Histochem Cytobiol 2009;47:65-8. [Crossref] [PubMed]
- van der Putten LJM, van Hoof R, Tops BBJ, et al. Molecular profiles of benign and (pre)malignant endometrial lesions. Carcinogenesis 2017;38:329-35. [Crossref] [PubMed]
- Kimmelman AC. Metabolic dependencies in RAS-driven cancers. Clin Cancer Res 2015;21:1828-34. [Crossref] [PubMed]
- Cox AD, Der CJ, Philips MR. Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res 2015;21:1819-27. [Crossref] [PubMed]
- Bouclier C, Simon M, Laconde G, et al. Stapled peptide targeting the CDK4/Cyclin D interface combined with Abemaciclib inhibits KRAS mutant lung cancer growth. Theranostics 2020;10:2008-28. [Crossref] [PubMed]
- McCormick F. KRAS as a therapeutic target. Clin Cancer Res 2015;21:1797-801. [Crossref] [PubMed]
- Ring KL, Yates MS, Schmandt R, et al. Endometrial cancers with activating KRas mutations have activated estrogen signaling and paradoxical response to MEK inhibition. Int J Gynecol Cancer 2017;27:854-62. [Crossref] [PubMed]
- Birkeland E, Wik E, Mj SS, et al. KRAS gene amplification and overexpression but not mutation associates with aggressive and metastatic endometrial cancer. Br J Cancer 2012;107:1997-2004. [Crossref] [PubMed]
- Shen L, Qu X, Ma Y, et al. Tumor suppressor NDRG2 tips the balance of oncogenic TGF-β via EMT inhibition in colorectal cancer. Oncogenesis 2014;3:e86. [Crossref] [PubMed]
- Yu J, Hua R, Zhang Y, et al. DNA hypomethylation promotes invasion and metastasis of gastric cancer cells by regulating the binding of SP1 to the CDCA3 promoter. J Cell Biochem 2020;121:142-51. [Crossref] [PubMed]
- Yue L, Li L, Liu F, et al. The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis 2013;34:927-35. [Crossref] [PubMed]
- Wang F, Li Y, Zhou J, et al. miR-375 Is Down-Regulated in Squamous Cervical Cancer and Inhibits Cell Migration and Invasion via Targeting Transcription Factor SP1. Am J Pathol 2011;179:2580-8. [Crossref] [PubMed]
- Wang S, Li Y, Sun S, et al. Sp1 promotes ovarian cancer cell migration through repressing miR-335 expression. Biochem Biophys Res Commun 2020;524:211-6. [Crossref] [PubMed]
- Zhao Y, Zhang W, Guo Z, et al. Inhibition of the transcription factor Sp1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol Rep 2013;30:1782-92. [Crossref] [PubMed]
- Shao W, Li Y, Chen F, et al. Long non-coding RNA DLEU1 contributes to the development of endometrial cancer by sponging miR-490 to regulate SP1 expression. Die Pharmazie 2018;73:379-85. [PubMed]
- Moskovitz J. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci U S A 2001;98:12920-5. [Crossref] [PubMed]
- Chen K, Liu H, Liu Z, et al. Genetic variants in RUNX3, AMD1 and MSRA in the methionine metabolic pathway and survival in nonsmall cell lung cancer patients. Int J Cancer 2019;145:621-31. [Crossref] [PubMed]
- Xu Y, Fulciniti M, Samur MK, et al. YWHAE 14-3-3ε expression impacts the protein load contributing to proteasome inhibitor sensitivity in multiple myeloma. Blood 2020;136:468-79. [Crossref] [PubMed]
- Oliva E, Loening T, Carcangiu ML, et al. Tumours of the uterine corpus: mesenchymal tumours. In Kurman RJ, Carcangiu ML, Herrington CS, et al. editors. WHO Classification of tumours of female reproductive organs. 4th ed. Lyon: IARC Press, 2014:135-47.
- Vajpeyi R. WHO classification of tumours: pathology and genetics of tumours of the breast and female genital organs. J Clin Pathol 2005;76:139-41.
- Cheah PS, Ramshaw HS, Thomas PQ, et al. Neurodevelopmental and neuropsychiatric behaviour defects arise from 14-3-3ζ deficiency. Mol Psychiatry 2012;17:451-66. [Crossref] [PubMed]
- Bjeije H, Soltani BM, Behmanesh M, et al. YWHAE long non-coding RNA competes with miR-323a-3p and miR-532-5p through activating K-Ras/Erk1/2 and PI3K/Akt signaling pathways in HCT116 cells. Hum Mol Genet 2019;28:3219-31. [Crossref] [PubMed]
- Oien DB, Osterhaus GL, Latif SA, et al. MsrA knockout mouse exhibits abnormal behavior and brain dopamine levels. Free Radic Biol Med 2008;45:193-200. [Crossref] [PubMed]
- Deng Y, Jiang B, Rankin CL, et al. Methionine sulfoxide reductase A (MsrA) mediates the ubiquitination of 14-3-3 protein isotypes in brain. Free Radic Biol Med 2018;129:600-7. [Crossref] [PubMed]
- Corso G, Pascale V, Flauti G, et al. Oncogenic mutations and microsatellite instability phenotype predict specific anatomical subsite in colorectal cancer patients. Eur J Hum Genet 2013;21:1383-8. [Crossref] [PubMed]
- Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 2008;359:1757-65. [Crossref] [PubMed]
- Lee CH, Ou WB, Marino-Enriquez A, et al. 14-3-3 fusion oncogenes in high-grade endometrial stromal sarcoma. Proc Natl Acad Sci U S A 2012;109:929-34. [Crossref] [PubMed]
- Gremel G, Liew M, Hamzei F, et al. A prognosis based classification of undifferentiated uterine sarcomas: Identification of mitotic index, hormone receptors and YWHAE-FAM22 translocation status as predictors of survival. Int J Cancer 2015;136:1608-18. [Crossref] [PubMed]


