
T-lymphoblastic lymphoma/leukemia without clonal TCR gene rearrangements: case report and literature review
Introduction
T-lymphoblastic lymphoma (T-LBL) is a highly aggressive malignancy originating from T-lymphocyte precursors. Incidence is highest in children and adolescents. Clinical onset is insidious, typically featuring painless cervical lymphadenopathy or mediastinal nodal enlargement that can cause tracheobronchial and esophageal stenosis, leading to dyspnea and dysphagia. By convention, the term lymphoma is used when the process is confined to a mass lesion with no or minimal evidence of peripheral blood and bone marrow involvement. With extensive bone marrow and peripheral blood involvement, the appropriate term is T-ALL. If a patient presents with a mass lesion and lymphoblasts in the bone marrow, the distinction between leukemia and lymphoma is arbitrary. In many treatment protocols, a value of >25% bone marrow blasts are used to define leukemia. Unlike with myeloid leukemias, there is no agreed upon lower limit for the proportion of blasts required to establish a diagnosis of ALL. In general, the diagnosis should be avoided when there are <20% blasts (1). CD3, CD7, and TDT-positive cells are the predominant immunophenotypes. T-cell receptor (TCR) gene rearrangement is usually present; TCR gene rearrangement-negative cases are considered rare. It is essential to differentiate T-LBL/T-ALL from other T-cell lymphoproliferative diseases. Detection of TCR gene rearrangement is highly sensitive and specific for the diagnosis of T-cell lymphoma. A study of TCR gene rearrangement in lymphoproliferative disorders found that 96.7% (29/30) of cases of T-cell lymphoma, including 100% (2/2) of cases of T-LBL/ALL, were TCR gene rearrangement-positive, while 100% (30/30) of cases of other T-cell lymphoproliferative diseases were TCR rearrangement-negative (2). However, the absence of TCR rearrangement does not exclude the diagnosis of T-cell lymphoma. Diagnosis should be based on clinical presentation, histopathology, immunotyping, and molecular genetics. We report a case of TCR gene rearrangement-negative T-LBL/ALL, discuss its clinical and pathological features, and review relevant literature. We present the following case in accordance with the CARE reporting checklist (available at http://dx.doi.org/10.21037/tcr-20-2902).
Case presentation
An 18-year-old male was hospitalized with a four-month history of cervical lymphadenopathy and the onset of dysphagia one month prior to admission. He denied fever, bone pain, or night sweats. Physical examination disclosed multiple palpable non-tender, matted and mobile cervical lymph nodes that were approximately 1.0–1.5 cm in diameter. Laboratory findings included WBC 8.62×109/L, with lymphoma cells accounting for 15% of peripheral blood leukocytes and an absolute value of 1.29×109/L; PLT 213×109/L; HGB 157 g/L; and LDH 307 U/L. Computed tomography revealed diffuse polyglandular lymphadenopathy of mediastinal, pelvic, retroperitoneal, and bilateral cervical, axillary, and inguinal nodes; splenomegaly with heterogenous radiodensity featuring areas of hypodensity; and pericardial thickening.
Histopathologic examination disclosed diffusely distributed small to medium sized round and oval cells with scant cytoplasm, delicate chromatin, and frequent mitotic figures. Immunophenotyping and ISH showed that tumor cells were positive for TDT, CD3, CD7, CD5, and CD10, with a ki-67 proliferation index of approximately 50%. MPO, CD1a, CD56, CD20, CD4, CD8, CD30, CD34, bcl-6, and EBER were negative (Figure 1). Molecular pathology assays for TCR gene rearrangements were negative (Table 1). Bone marrow aspirate disclosed lymphoblasts that accounted for 58.88% cells. Flow cytometry revealed that all lymphoblasts expressed CD3, CD5, CD7; partial expression of CD2, CD10, CD38, TDT; and no expression of CD1a, CD34, CD4, CD8, mCD3, CD33, CD56, CD19, CD79a, HLA-DR and MPO. These findings led to the diagnosis of T-LBL/-ALL. The patient received chemotherapy consisting of vinorelbine, pirarubicin, cyclophosphamide, asparaginase, and prednisone. Prophylactic chemotherapy of the central nervous system and radiotherapy of the mediastinum were also given. Four months later, he was in clinical remission. Bone marrow and cerebrospinal fluid analyses yielded normal results, indicating a complete response.
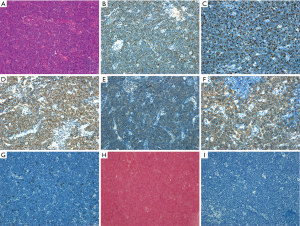
Table 1
| Test item | Gene rearrangement type | Result |
|---|---|---|
| T lymphocyte gene rearrangement test | TCRB A tube (Vβ+Jβ1/2) | Negative |
| TCRB B tube (Vβ+Jβ2) | Negative | |
| TCRB C tube (Dβ+Jβ1/2) | Negative | |
| TCRG A tube (Vγ1-8, Vγ10) | Negative | |
| TCRG B tube (Vγ9, Vγ11) | Negative |
*Detailed test diagrams are available in supplementary information.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from individual participants.
Discussion
T-LBL is a clonal hematopoietic stem cell malignancy characterized by an immature T-cell phenotype. Most T-LBL originates in the mediastinum, probably because the thymus is the primary site of T-cell maturation (3). Cytologic findings are relatively consistent, featuring small to medium sized cells with scant cytoplasm, although some cases may exhibit a large cell morphology. Other features include round nuclei, delicate chromatin, and small and unclear nucleoli. Most T-LBL/ALL cells express TDT (94.5%), CD7 (96.3%) and CD3 (72.3%), Cytoplasmic CD3 is thought to be the most specific marker of T-cell lineage, but T-lymphoblasts can express other markers related to T-cell differentiation and maturation in various combinations. The expression of CD2 may be related to prognosis (3-5).
The clinical features, histopathology, and immunohistochemical, and bone marrow flow cytometric findings of this case are consistent with T-LBL/ALL. TCR gene rearrangement is an important diagnostic finding in T-LBL/ALL. Almost all cases exhibit clonal TCR rearrangement, and approximately 20% of cases are accompanied by IGH gene rearrangement (1). Because TCR gene rearrangement-negative T-LBL/ALL cases are relatively rare, and because TCR gene rearrangement-negativity does not exclude the diagnosis of T-LBL/ALL, statistics regarding the incidence and prevalence of TCR gene rearrangement-negative T-LBL/ALL are not available. Consequently, clonal TCR gene rearrangement-positivity can support the diagnosis of T-LBL, but a negative result does not exclude the diagnosis. Although a 2005 study reported by Lin and colleagues (6) proposed that some T-LBL may be NK-LBL, the 2017 edition of the World Health Organization (WHO) classification standard for hematologic malignancies (1) clearly pointed out that it has been difficult to define NK-ALL/LBL, which is quite confusing in the literature. NK precursor cells do not express specific markers or overlap with the markers of T-ALL/LBL in the early stage of development. According to the 2017 WHO classification standard (1), CD56-negative tumors are categorized as T-LBL/ALL. All subjects reported by Lin (6) tested negative for CD56. Consequently, the NK-LBL discussed in their study should be classified as T-LBL/ALL. In our case and in previous studies (4-6), 14 patients with TCR gene rearrangement-negative T-LBL/ALL have been reported (Table 2). With the exception of one case that did not undergo immunohistochemistry, all were CD3 and TDT positive and EBER negative, indicating immature T-cells without Epstein-Barr virus-mediated oncogenesis. In this case and in a report by Li and colleagues (5), three patients were positive for CD7, an early T cell marker that indicates that T-cell precursors are the progenitors of tumor cells. Under normal conditions, early T-cell precursors first enter the thymic cortico-medullary junction and then migrate to the outer cortical region, acquiring the CD5 and CD1 phenotypes and losing the HLA-DR antigen. The development of double negative T-cells that do not express CD4 and CD8; followed by a rearrangement of TCR gene sequences of the δ, γ, β, and α lines; followed by positive and negative selection in the thymus; results in cell populations with functional TCRs (2). However, the molecular genetic results of this case identified no clonal TCR gene rearrangement, and showed that tumor cell differentiation was arrested at the early T-cell precursor stage. The pathogenesis of lymphoma is unclear and potentially multifactorial. Etiologies may include genetic factors, oncogenic infections, physical radiation, and chemical carcinogens. A growing number of studies have found that many lymphomas are associated with mutations and other molecular genetic abnormalities. The NOTCH1 signaling pathway has an important regulatory function in TCR assembly and signal transduction in early and immature T-cells, and is the most studied pathway of T-LBL/ALL oncogenesis. Hounjet and colleagues (7) found that >60% of T-LBL/ALL cases exhibited oncogenic NOTCH1 mutations. Consequently, the NOTCH-1 signaling pathway has emerged as a potential therapeutic target. The clinical development of small moleculeγ-secretase inhibitors (GSI) that target the NOTCH1 signaling pathway has been hindered by dose-limiting toxicity. However, the anti-malarial drug chloroquine (CQ) inhibits lysosomal function and autophagy; interferes with intracellular transport; reduces the activity and proliferation of T-ALL cells; antagonizes NOTCH1 signaling; and exerts synergy withγ-secretase inhibition. Consequently, CQ may be a promising drug for the treatment of T-ALL, either as monotherapy or in combination with GSI (7). Lin and colleagues (6) proposed that because natural killer (NK) and T-cells are derived from a common precursor in the thymus, some cases with clinical and histologic features of T-LBL may arise from immature NK cells. They identified seven cases of CD3- and TDT-positive/TCR gene rearrangement-negative LBL expressing the NK cell marker CD94 1A, suggesting an NK cell lineage, and leading to their suggestion of a new diagnostic category, NK-LBL. These patients experienced significantly improved two-year survival compared to patients with TCR rearrangements (100% vs. 27%, P<0.01). All of the NK-ALL/LBL patients reported by Lin (6) belong to T-ALL/LBL according to the 2017 WHO classification (1), because of negative CD56 expression. Are there cases of CD56-negative NK-ALL/LBL? Will CD56-negative NK-ALL/LBL carry a better prognosis than T-ALL/LBL? More research is needed to address these issues, but until conclusive evidence emerges, CD56-negative cases are classified as T-ALL/LBL (1).
Table 2
| Case | Age | Sex | EBER | CD3 | CD4 | CD56 | CD7 | CD8 | TDT | CD20 | CD10 | CD34 | Bcl-6 | MPO | Ki67% | TCR | Therapy* | Mo# | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 18 | M | − | + | − | − | + | − | + | − | + | − | − | − | 50% | − | No | 4+ | Present |
| 2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | − | NA | NA | NA | − | NA | − | NA | NA | (3) |
| 3 | 73 | M | − | + | + | + | + | − | + | − | − | + | + | − | 50% | − | No | 5+ | (4) |
| 4 | 73 | M | − | + | + | − | + | + | + | − | + | + | + | − | 60% | − | No | 9+ | (4) |
| 5 | 52 | F | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | No | 30 | (5) |
| 6 | 53 | F | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | No | 120+ | (5) |
| 7 | 29 | F | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | No | 14 | (5) |
| 8 | 3 | M | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | Yes | 34+ | (5) |
| 9 | 19 | F | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | No | 78+ | (5) |
| 10 | 12 | F | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | Yes | 105+ | (5) |
| 11 | 27 | M | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | No | 108+ | (5) |
| 12 | 12 | M | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | Yes | 108+ | (5) |
| 13 | 15 | M | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | Yes | 50+ | (5) |
| 14 | 28 | F | − | + | NA | − | NA | NA | + | − | NA | NA | NA | NA | NA | − | No | 45+ | (5) |
*Patients were treated with either ALL-like or CHOP (cyclophosphamide/doxorubicin/vincristine/prednisone) regimens. #The time unit of observation is expressed in months (Mo).
The differential diagnosis of T-LBL includes: (I) Mantle cell lymphoma mother cell variant that can be differentiated by TDT-negative and CD20, Cyclin D1, and CD5-positive findings; (II) small round blue cell tumors (including Ewing's sarcoma and neuroblastoma) that can be identified by adherent growth without expression of lymphocyte markers; (III) B-lymphoblastic lymphoma, that can be identified by CD79a and PAX5 expression, in contrast to CD3 and CD7 expression in T-LBL; (IV) acute myeloid leukemia (AML); both T-LBL and AML can express TDT, CD3, CD4, and CD43, but expression intensities of AML are weaker than that of T-LBL, and AML usually expresses MPO lysozyme and CD117; (V) indolent T-lymphoblastic proliferative lesions; these typically involve the upper respiratory and digestive tracts, and are characterized by multi-site recurrence, but without systemic involvement; these lesions are morphologically and immunophenotypically indistinguishable from T-LBL/ALL, but lacking clonal rearrangement of TCR genes (8); (VI) NK-LBL/ALL; this can be diagnosed by blastocytic cell morphology; expression of TDT; expression of CD56 and related markers of immature T cells such as CD2, CD7, cytoplasmic CD3, etc.; the absence of B-cell and myeloid markers; the absence of IGH and TCR gene rearrangement; and exclusion of blastocytic cell plasmacytoid dendritic cell tumor (1).
Although the detection of TCR gene rearrangement can facilitate the diagnosis of T-LBL/ALL, there are no specific therapeutic regimens for T-LBL/ALL based on TCR gene rearrangement-positivity or negativity. Both TCR rearrangement positive- and negative-T-LBL/ALL are treated with similar multi-drug high-dose chemotherapy regimens such as Hyper-CVAD (cyclophosphamide, Changchun sheen, dexamethasone, and epirubicin); or BFM-90; combined with central nervous system prophylaxis and local radiotherapy. The addition of autologous stem cell transplantation or bone marrow transplantation significantly improves complete response and overall survival rates (9). Prognostic factors include age, gender, size of the primary mass, mediastinal and bone marrow involvement, peripheral blood tumor cell and LDH contents, Ann Arbor stage, and treatment regimen. Our patient benefited from the following factors: on one hand, his young age and timely detection and diagnosis; on the other hand, the positive combination of high dose chemoradiotherapy and central nervous system prophylaxis that achieved relatively good clinical efficacy.
In conclusion, T-LBL/ALL with negative TCR gene rearrangement is rare, and should be carefully differentiated from diseases such as indolent T-cell lymphoblastic proliferative lesions and NK-LBL/ALL. Diagnosis is facilitated by assessing clinical findings, histopathology, immunohistochemistry, and molecular genetics.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at http://dx.doi.org/10.21037/tcr-20-2902
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tcr-20-2902). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues: Revised Fourth Edition, 2017.
- Pan XY, Yang CS, Li GY, et al. Analysis of TCR gene rearrangements for diagnosis of T-cell lymphomas. Chinese J Clin Exp Pathol 2015;4:400-3.
- Jaffe ES, Arber DA, Campo E, et al. Hematopathology, Second Edition. Amsterdam: Elsevier, 2017.
- Yu B, Du JR, Xie JL, et al. Clinicopathologic study of 128 cases of T-lymphoblastic lymphoma/leukemia. Zhonghua Bing Li Xue Za Zhi 2010;39:452-7. [PubMed]
- Li X, Wang Y, Chen R, et al. Clinicopathologic features and prognosis of T lymphoblastic lymphoma associated with Langerhans cell histiocytosis. Zhonghua Bing Li Xue Za Zhi 2014;43:522-7. [PubMed]
- Lin CW, Liu TY, Chen SU, et al. CD94 1A transcripts characterize lymphoblastic lymphoma/leukemia of immature natural killer cell origin with distinct clinical features. Blood 2005;106:3567-74. [Crossref] [PubMed]
- Hounjet J, Habets R, Schaaf MB, et al. The anti-malarial drug chloroquine sensitizes oncogenic NOTCH1 driven human T-ALL to gamma-secretase inhibition. Oncogene 2019;38:5457-68. [Crossref] [PubMed]
- Ohgami RS, Arber DA, Zehnder JL, et al. Indolent T-lymphoblastic proliferation (iT-LBP): a review of clinical and pathologic features and distinction from malignant T-lymphoblastic lymphoma. Adv Anat Pathol 2013;20:137-40. [Crossref] [PubMed]
- Hu M, Wang H, Wang L, et al. Outcome of adult T-lymphoblastic lymphoma depends on ALL-type chemotherapy, prognostic factors, and performance of allogeneic hematopoietic stem cell transplantation. Medicine (Baltimore) 2018;97:e11374. [Crossref] [PubMed]


