
Low expression of RalGAPs associates with the poorer overall survival of head and neck squamous cell carcinoma
Introduction
Head and neck carcinoma mainly refers to a group of malignancies that derives from the moist mucosal surfaces lining oral cavity, pharynx (including nasopharynx, oropharynx, and hypopharynx), larynx, paranasal sinuses, and nasal cavity. Head and neck squamous cell carcinoma (HNSC) begins in the squamous cells of mucosa and accounts for more than 90% of all head and neck carcinomas and contributes to high lethality (1,2). HNSC is estimated to cause over 600,000 new cases and 380,000 deaths yearly worldwide (3,4).
Ras like (Ral) small GTPases belong to the Ras branch of the Ras superfamily and consists of two proteins, RalA and RalB, sharing 82% identity at the amino acid level (5). Accumulating evidence indicates that Ral small GTPases play an important role in tumorigenesis and development of human cancers, including medulloblastoma (6), hepatocellular carcinoma (7), breast (8), colorectal (9), and lung cancer (10). Owing to the high affinity of Ral small GTPases for guanidine nucleotides, the complex protein-protein interactions facilitating Ral mediated signaling cascades, and the “smooth” surface of activated Ral GTP proteins, it is very difficult to produce small molecule compounds that can effectively and directly inhibit Ral activity (5). Moreover, Ral mutations are rare in human cancers, enabling the development of indirect inhibitors targeting the Ral regulators (5). Ral works with other small GTPases to transform the inactive GDP form to the active GTP form and vice versa, where it is hydrolyzed by GTPase-activating proteins (RalGAPs) and catalyzed by guanine nucleotide exchange factors (RalGEFs). To date, seven RalGEFs (RalGDS, RalGPS1, RalGPS2, RGL1, RGL2, RGL3, and RGL4) and two RalGAP heterodimers (catalytic subunit RalGAPα1, RalGAPα2, and the common regulatory subunit RalGAPβ) have been described in the literature (5,11). Vigil et al. have reported that RGL2 (a RalGEF) overexpression enhances pancreatic ductal adenocarcinoma (PDAC) growth via Ral activation in a Ral-independent manner (12). Conversely, the loss of RalGAP promotes the progression of bladder cancer (13) and enhances local invasion of prostatic intraepithelial neoplasia (14). However, the role of Ral and RalGAPs on the progression of HNSC remains unclear.
Methylation of the cytosine residue at position C5 within CpG dinucleotides is a known epigenetic modification in mammals, which has been strongly associated with the development of various cancers (15). If a hypermethylated area is changed to a hypomethylated area within the repeat-rich pericentromeric heterochromatin, mitotic recombination and genomic instability occur, resulting in cancer. If CpG islands of tumor suppressor gene (TSG) are hypermethylated, TSG will lose its ability to inhibit cancer (15). The hypermethylation of cycle genes, apoptosis-related genes, genes of the Wnt signaling pathway, cell adhesion molecules, DNA repair genes, and TSGs has been linked to the early events and advanced stages of HNSC (16). However, the relationship between DNA methylation and RalGAP expression on HNSC is rarely reported.
In this study, we demonstrated that downregulation of the RalGAP subunit could activate RalA. RalA and RalGAPs could act as predictive biomarker to evaluate prognosis of HNSC. And DNA methylation might be a potential mechanism for the downregulated expression of RalGAPα2. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tcr-21-1489).
Methods
Cell culture
The human OSCC cell lines, HSC-2, HSC-3, OSC20 and TSU were obtained and cultured as previously described (17). Briefly, cells were cultivated in high glucose Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37 °C in a humidified incubator with 5% CO2.
Small interfering RNA treatment
Cells were transfected with three different predesigned small interfering RNAs (siRNAs, 10 nM; Ambion, Thermo Fisher Scientific, Waltham, MA, USA) for each gene (RalGAPα1, RalGAPα2 and RalGAPβ) using Lipofectamine RNAiMAX (Invitrogen, Waltham, MA, USA) according to the manufacturers’ instructions. Cells transfected with an optimal siRNA for 48 h were used for experiments.
GST-Sec5 pull down assay
Glutathione-S-transferase (GST) fusion protein, which includes the Ral-binding domain from Sec5, and is known to be able to associate with GTP-bound active Ral, was synthesized and purified as previously described (18). After serum starvation for 6 h, cells were stimulated using medium supplemented with 10% FBS for 5 min at 37 °C. The cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed with the lysis buffer [50 mM HEPES/KOH, pH 7.4, 100 mM NaCl, 4 mM MgCl2, 2 mM EGTA, 1 mM dithiothreitol, 0.1% (w/v) Triton X-100, 10 mM b-glycerophosphate, 10 mM NaF, 1 mM Na3VO4, and EDTA-free protease inhibitor cocktail (Roche)]. After shaking at 4 °C for 15 min, the cell lysates were centrifuged at 100,000 ×g for 10 min. Glutathione Sepharose (GE Healthcare, USA) coated with 20 μg GST-Sec5 fusion protein was incubated with cell lysate containing 200 μg protein and rotated at 4 °C for 1 h. After washing 3 times in lysis buffer, the associated proteins were eluted using elution buffer (50 mM Hepes 7.4, 100 mM KCl, 1 mM DDT, 20 mM Glutathione of reduced form) at 4 °C for 30 min. The eluted supernatants were then subjected to western blot.
Western blot
Proteins were transferred to a PVDF membrane (Wako, Japan) after electrophoresis using a 12.5% e-PAGE gel (ATTO, Japan). Five percent skim milk was used to block nonspecific antigens and then the membranes were incubated with the appropriate primary antibody (1:1,000) at 4 °C overnight. After incubation with horseradish peroxidase-conjugated secondary antibodies, the blots were visualized. The antibodies used in this analysis were described in Table S1.
Bioinformatic analysis
The data of TCGA and GTEx were combined to analyze the pan-cancer gene expression of RalA and RalGAPs in cancer and adjacent normal tissues (https://sangerbox.com/), and the mutation status of RalGAPs. The relationships between overall survival of HNSC patients (n=488) and high or low expression of RalA or RalGAPs were evaluated by RNA sequencing analysis (19). The data were adopted from the Kaplan-Meier plotter (https://kmplot.com/analysis/).
Methylation-specific PCR (MSP)
A total of 1×105 cells were collected from each sample and subjected to DNA isolation and bisulfite treatment using the EpiTect Plus DNA Bisulfite Kit (QIAGEN, Germany) following the manufacturer’s instructions. Specific primers, listed in Table S2, were designed by MethPrimer (https://www.urogene.org/methprimer2/) software (20). Primer pairs targeting the methylated (M) or unmethylated (U) promoter regions (CpG islands) ofRalGAPα1 and RalGAPα2 were used for MSP. The converted DNA was PCR-amplified using an EpiTaq HS kit (TaKaRa, Japan) and the PCR amplification conditions were as follows: 35 cycles of 98 °C for 10s, 55 °C for 30 s, 72 °C for 30 s. PCR products were then electrophoresed using a 1% agarose gel containing ethidium bromide.
Next-generation sequencing-bisulfite sequencing PCR (NGS-BSP) analysis
The gene-specific DNA methylation patterns from normal epithelium and paired cancer tissues from six patients with HNSC were assessed using NGS-BSP which was completed using a previously published method (17,21,22). Briefly, BSP primers were designed using online MethPrimer software and were listed in Table S2. Genomic DNA (1 μg) was converted to template using ZYMO EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA), and one twentieth of the elution products were used as templates for the 35 cycle PCR amplifications completed using KAPA 2G Robust HotStart PCR Kit (Kapa Biosystems, Wilmington, MA, USA). The BSP products of multiple genes were then pooled, 5'-phosphorylated, 3'-dA-tailed and ligated to barcoded adapters using T4 DNA ligase (New England Biolabs, Ipswich, MA, USA). Barcoded libraries from each of the samples were then sequenced using an Illumina platform. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of the West China Hospital of Stomatology, Sichuan University (No. WCHSIRB-ST-2016-153) and informed consent was taken from all the patients.
Statistical analysis
Data were shown as the mean ± standard error of mean (S.E.M.) from 3 independent experiments with biological replicates. Student’s t-test was used for comparisons between two groups. Overall survival was evaluated using a Kaplan-Meier analysis, and the significance was calculated using log-rank test. Wilcoxon test was used to compare the methylation status of HNSC and paired adjacent normal epithelial tissues. A P value of <0.05 was considered statistically significant.
Results
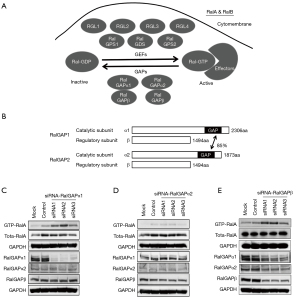
RalGAPs expression was negatively associated with RalA activation
RalGEFs consist of 7 proteins (RalGDS, RalGPS1, RalGPS2, RGL1, RGL2, RGL3, and RGL4) and catalyze active GDP-Ral to inactive GTP-Ral (Figure 1A). RalGAPs are heterodimeric complexes and consist of 3 different subunits (RalGAPα1, RalGAPα2, and the common RalGAPβ subunit), hydrolyzing active GTP-Ral to inactive GDP-Ral (Figure 1A). The RalGAPα1 and RalGAPα2 subunits act as catalytic function, nevertheless the RalGAPβ act as regulatory role. In mammals, α1 and α2 are highly identical at overall amino acid level overall (Figure 1B). As previously described, the expression of RalGAPs in eight HNSC cell lines was distinct, and those with higher level of RalGAPs tended to have lower level of GTP-RalA (17). Therefore, transfection of siRNA-RalGAPα1 into HSC-3 cells, which highly expressed endogenous RalGAPα1, dramatically reduced RalGAPα1 expression and increased RalA activation (Figure 1C). However, siRNA-RalGAPα2 did not apparently knock down the expression of RalGAPα2 and activate RalA (Figure 1D). Knock-down of siRNA-RalGAPβ decreased all RalGAPs expression but enhanced RalA activation since RalGAPβ is indispensable for the stability of RalGAP heterodimers (Figure 1E).
HNSC patients with high RalA expression or low RalGAP expression had worse overall survival (Figure 2)
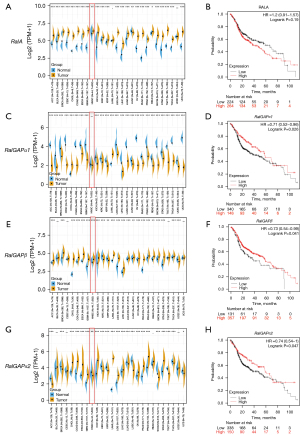
The combined analysis of TCGA and GTEx database showed that the expression of RalA in HNSC tissues was significantly higher than that in the adjacent normal tissues (P<0.01; Figure 2A). The Kaplan-Meier plotter data (19) of 488 patients with HNSC manifested that those patients with high level of RalA tended to have poorer prognosis, although there was no statistical difference (Figure 2B). Although the difference of RalGAPα1 in cancer and normal tissues was not significant (Figure 2C) and RalGAPβ was higher in cancer tissues (Figure 2E), HNSC patients with low RalGAPα1 and RalGAPβ expression had worse overall survival (logrank P<0.05; Figure 2D,2F). Conversely, the expression of RalGAPα2 in HNSC tissues was significantly lower than that in the adjacent normal tissues (P<0.01; Figure 2G), and Kaplan-Meier demonstrated that HNSC patients with low RalGAPα2 expression had worse overall survival (logrank P<0.05; Figure 2H).
RalGAPα2 promoter was hypermethylated in vitro and in vivo, associating with lymph node metastasis
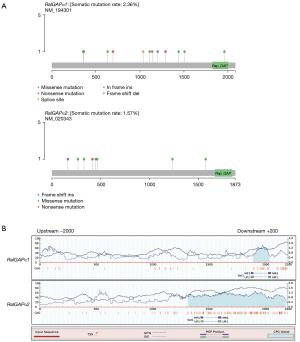
Given the low-expressed tendency of core catalytic subunits, RalGAPα1 and RalGAPα2, in HNSC tissues, we examined their mutation status in HNSC TCGA database, and we found that the somatic mutations of RalGAPα1 and RalGAPα2 were very low (Figure 3A). DNA methylation is associated with gene silencing (23), thus we hypothesized that DNA methylation might be involved in the suppression of RalGAPα2. We focused on the RalGAPα2 promoter from 2,000 bp upstream of transcription start site (TSS) to 200 bp downstream of the TSS. CpG island prediction and primer design were completed using Methprimer tool (Figure 3B) and MSP was used to screen the methylation status in four HNSC cell lines. The RalGAPα2 promoter was widely methylated in comparison with RalGAPα1 (Figure 4A).
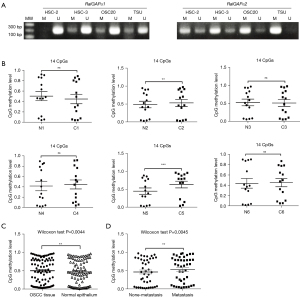
NGS-BSP analysis of six samples from patients with HNSC showed that the 14 CpG loci within the RalGAPα2 promoter in cancer tissues exhibited a tendency to be highly methylated than in paired-adjacent normal epithelium samples, although only two pairs showed statistical significance (Figure 4B). The difference in total methylation levels for all 14 CpG loci was statistically significant (Wilcoxon test P=0.0044; Figure 4C) in each paired tissue. These six patients were then divided into two groups, with (n=3) or without (n=3) neck lymph node metastasis. The total methylation levels of patients with neck lymph node metastasis were significantly higher than that in non-metastasis group (Wilcoxon test P<0.05; Figure 4D).
Discussion
The RAS gene, is the most common oncogene family and comprises three genes (HRAS, NRAS, and KRAS), which are completely mutated in more than one-third of human cancers (24). As one of the subfamilies of Ras proteins, Ral GTPases can be activated by mutated RAS (25). Ral is one of the downstream effectors of Ras and works in combination with the other well-known Ras partners RAF-MAPK-ERK and PI3K-AKT-mTOR (5). We previously described several RAS mutations in the eight HNSC lines, and none of these mutations, with the exception of the G12S active mutation in the HRAS gene of HOC313 cells, were found to be functionally important (17), which is consistent with the low rate of RAS mutations in HNSC, as described in the COSMIC v92 database (https://cancer.sanger.ac.uk/cosmic). Thus, other regulatory mechanisms, may be primarily responsible for the aberrant activation of Ral in HNSC.
Like Ras and other GTPases, RalGTPases cycle between active GTP-bound and inactive GDP-bound forms (11). RalGEFs promote the production of Ral-GTP by catalyzing the exchange of GDP and GTP, while RalGAPs hydrolyze GTP to GDP, restoring Ral to its inactive state (5). Due to the high degree of structural similarity between RalA and RalB it is unsurprising that they present with identical enzymology and are modulated by the same series of RalGEFs and RalGAPs (5,11). However, RalA and RalB have different effects on cellular function and the progression of human cancers, partly resulting from the differences in their C-terminal sequences, which mediate differences in their subcellular membrane localizations as well as their effector usage (26). In PDAC, RalA has been shown to be involved in anchorage-independence and tumorigenic growth, while RalB is required for Matrigel invasion and lung metastasis (27). In colorectal cancer (CRC), RalA and RalB exert antagonistic functions on anchorage-independent growth, with RalA inhibition attenuating anchorage-independent growth, while RalB suppression promoted soft agar colony formation (28). These strikingly different roles in various cancers are the result of the differences in RalA and RalB effectors. RalA-Exo84 interactions are required for anchorage-independent growth while RalB-Sec5 interactions inhibit anchorage-independent growth (28). Intriguingly, both PADC and CRC have a high rate of RAS mutation (https://cancer.sanger.ac.uk/cosmic).
We uncovered a high degree of similarity in RalA and RalB activation in eight HNSC cell lines in a previous study (17). In this study, we focused on RalA activation, which is mainly associated with downregulation of RalGAPs. We demonstrated that RalA activation was largely dependent on the downregulation of RalGAP subunits (Figure 1C,1E). The reason why siRNA-RalGAPα2 did not apparently knock down the expression of RalGAPα2 and activate RalA may account for the extremely low endogenous expression of RalGAPα2 in HSC-3 cells, as previously described (17). The expression of RalA in HNSC tissues was significantly higher than in the adjacent normal tissues. And HNSC patients with high level of RalA tended to have poorer prognosis (Figure 2B). Conversely, the expression of RalGAPα2 in HNSC tissues was significantly lower than that in the adjacent normal tissues, and HNSC patients with low RalGAPα2 expression had worse overall survival. Previous studies also demonstrated that reduced RalGAPα2 expression was linked to reduced overall survival in bladder (13) and prostate cancers (14), which was consistent with our finding that reduced expression of RalGAPα2 was associated with worse overall survival of HNSC (Figure 2F). These findings were consistent with our previously described conclusion that RalA activation in oral cancer cell lines was associated with downregulation of RalGAPs (17).
DNA methylation in promoters or regulatory elements is commonly associated with gene silencing or transcriptional repression via inhibiting transcription factor interactions or the condensation of the chromatin via histone deacetylase activity (29). In this study, we uncovered that the promoter of RalGAPα2 was hypermethylated in four HNSC cell lines when compared with the RalGAPα1 promoter (Figure 4A). The DNA methylation status of the RalGAPα2 promoter was higher in HNSC tissues than in their paired normal epithelia (Figure 4B,4C) and positively associated with neck lymph node metastasis in vivo (Figure 4D).
Conclusions
Our study demonstrated that RalA and RalGAPs could act as specific predictors to evaluate the prognosis of HNSC. DNA methylation might be a potential mechanism that downregulated the RalGAPα2 expression in HNSC. Accumulating evidence has revealed the negative regulatory role of RalGAPα2, suggesting that this gene is a tumor suppressor. Further understanding of this gene and its regulatory network may provide evidence supporting the application ofRalGAPα2 as a predictive biomarker and may reveal that this gene could be a therapeutic target in a variety of human cancers.
Acknowledgments
Funding: This study was supported by the Sichuan University West China Hospital of Stomatology Fund for Basic and Applied Basic Research (No. RD-02-201914) for PG, and Doctoral Scientific Project of the Natural Science Foundation of Shandong Province (No. ZR2018BH026) for XM.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tcr-21-1489
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tcr-21-1489
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tcr-21-1489). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committee of the West China Hospital of Stomatology, Sichuan University (No. WCHSIRB-ST-2016-153) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011;11:9-22. [Crossref] [PubMed]
- Yan K, Agrawal N, Gooi Z. Head and Neck Masses. Med Clin North Am 2018;102:1013-25. [Crossref] [PubMed]
- Global Burden of Disease Cancer Collaboration. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2017;3:524-48. [Crossref] [PubMed]
- Miyauchi S, Kim SS, Pang J, et al. Immune Modulation of Head and Neck Squamous Cell Carcinoma and the Tumor Microenvironment by Conventional Therapeutics. Clin Cancer Res 2019;25:4211-23. [Crossref] [PubMed]
- Hayes RB, Ahn J, Fan X, et al. Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer. JAMA Oncol 2018;4:358-65. [Crossref] [PubMed]
- Ginn KF, Fangman B, Terai K, et al. RalA is overactivated in medulloblastoma. J Neurooncol 2016;130:99-110. [Crossref] [PubMed]
- Ezzeldin M, Borrego-Diaz E, Taha M, et al. RalA signaling pathway as a therapeutic target in hepatocellular carcinoma (HCC). Mol Oncol 2014;8:1043-53. [Crossref] [PubMed]
- Zago G, Veith I, Singh MK, et al. RalB directly triggers invasion downstream Ras by mobilizing the Wave complex. Elife 2018;7:40474. [Crossref] [PubMed]
- Győrffy B, Stelniec-Klotz I, Sigler C, et al. Effects of RAL signal transduction in KRAS- and BRAF-mutated cells and prognostic potential of the RAL signature in colorectal cancer. Oncotarget 2015;6:13334-46. [Crossref] [PubMed]
- Biondini M, Duclos G, Meyer-Schaller N, et al. RalB regulates contractility-driven cancer dissemination upon TGFβ stimulation via the RhoGEF GEF-H1. Sci Rep 2015;5:11759. [Crossref] [PubMed]
- Shirakawa R, Horiuchi H. Ral GTPases: crucial mediators of exocytosis and tumourigenesis. J Biochem 2015;157:285-99. [Crossref] [PubMed]
- Vigil D, Martin TD, Williams F, et al. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J Biol Chem 2010;285:34729-40. [Crossref] [PubMed]
- Saito R, Shirakawa R, Nishiyama H, et al. Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene 2013;32:894-902. [Crossref] [PubMed]
- Uegaki M, Kita Y, Shirakawa R, et al. Downregulation of RalGTPase-activating protein promotes invasion of prostatic epithelial cells and progression from intraepithelial neoplasia to cancer during prostate carcinogenesis. Carcinogenesis 2019;40:1535-44. [Crossref] [PubMed]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet 2005;6:597-610. [Crossref] [PubMed]
- D'Souza W, Saranath D. Clinical implications of epigenetic regulation in oral cancer. Oral Oncol 2015;51:1061-8. [Crossref] [PubMed]
- Gao P, Liu S, Yoshida R, et al. Ral GTPase Activation by Downregulation of RalGAP Enhances Oral Squamous Cell Carcinoma Progression. J Dent Res 2019;98:1011-9. [Crossref] [PubMed]
- Shirakawa R, Fukai S, Kawato M, et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem 2009;284:21580-8. [Crossref] [PubMed]
- Nagy Á, Lánczky A, Menyhárt O, et al. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 2018;8:9227. [Crossref] [PubMed]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002;18:1427-31. [Crossref] [PubMed]
- Gao F, Liang H, Lu H, et al. Global analysis of DNA methylation in hepatocellular carcinoma by a liquid hybridization capture-based bisulfite sequencing approach. Clin Epigenetics 2015;7:86. [Crossref] [PubMed]
- Gao F, Xia Y, Wang J, et al. Integrated analyses of DNA methylation and hydroxymethylation reveal tumor suppressive roles of ECM1, ATF5, and EOMES in human hepatocellular carcinoma. Genome Biol 2014;15:533. [Crossref] [PubMed]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell 2012;150:12-27. [Crossref] [PubMed]
- Li S, Balmain A, Counter CM. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer 2018;18:767-77. [Crossref] [PubMed]
- Smith SC, Oxford G, Baras AS, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res 2007;13:3803-13. [Crossref] [PubMed]
- Martin TD, Der CJ. Differential involvement of RalA and RalB in colorectal cancer. Small GTPases 2012;3:126-30. [Crossref] [PubMed]
- Lim KH, O'Hayer K, Adam SJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol 2006;16:2385-94. [Crossref] [PubMed]
- Martin TD, Samuel JC, Routh ED, et al. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res 2011;71:206-15. [Crossref] [PubMed]
- Urbano A, Smith J, Weeks RJ, et al. Gene-Specific Targeting of DNA Methylation in the Mammalian Genome. Cancers (Basel) 2019;11:1515. [Crossref] [PubMed]


