
Identification of potential key molecular biomarkers in lung adenocarcinoma by bioinformatics analysis
Introduction
Lung cancer is one of the most common malignant tumors in the world, with recent statistics indicating that about 26% of tumor patients die from this disease (1). Moreover, lung adenocarcinoma (LUAD) is one of the most common pathological types of non-small cell lung cancer (NSCLC). In China, the incidence of lung cancer continues to follow an annually increasing trend (2), and LUAD has become the most common type. Therefore, it is necessary to explore the mechanism of occurrence and development of LUAD, particularly the abnormally expressed genes that play an important role. By discovering genes and studying their corresponding functions, it may be possible to provide new strategies for the treatment of LUAD.
Microarray technology and bioinformatics have been extensively used in tumor research. Many current studies are performed using various analytical tools to compare microarray datasets to obtain DEGs in various tumors. Then, the role of DEGs is explored in molecular functions (MFs), biological processes (BPs), and different pathways to provide new ideas for tumor research (3,4). However, due to various factors such as sample size, tumor stage, grade, and ethnicity, a complete set of DEGs is often not available. Therefore, it is important for us to compare the latest microarray datasets repeatedly in order to obtain more representative DEGs. Herein, 3 LUAD messenger RNA (mRNA) datasets were screened from the Gene Expression Omnibus (GEO) database, and a total of 284 DEGs were discovered by intersection. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were employed for the functional annotation of DEGs, and a protein-protein interaction (PPI) network was established through Cytoscape (https://cytoscape.org/). In summary, we used the above methods to illustrate the molecular mechanism of LUAD. As a result, 284 DEGs and 10 hub genes were screened, which may be used as potential biomarkers for LUAD. In this study, we found low expression of fibroblast growth factor 2 (FGF2) in LUAD and it is correlated with the long-term prognosis among the patients. Specifically, with higher FGF2 expression, patients tend to have a longer life span after diagnosis, which is not consistent with previous studies (5,6). Thus, more effort is needed to clarify the mechanism between FGF2 and LUAD. Herein, we explore the role of FGF2 in the immune infiltration of LUAD, which may be another prognostic factor that provides a novel prospect for the LUAD’s treatment. We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2676/rc).
Methods
Source of data
The datasets were collected from the open GEO (https://www.ncbi.nlm.nih.gov/geo/). We selected 3 lung cancer datasets, GSE2514 (7), GSE7670 (8), and GSE40275 (9), and human LUAD was the main pathological type. Among them, GSE2514 contains 19 tumor samples and 19 normal lung tissue samples, GSE7670 contains 57 tumor samples and 57 normal lung tissue samples, and GSE40275 contains 8 tumor samples and 43 normal lung tissue samples. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
DEGs screened
The GEO2R online tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to screen tumor tissues and normal human lung tissues, and statistically analyze each dataset to calculate the adjusted P value and |logFC| and define the required DEGs that meet the conditions, that is, adjusted P<0.01, |logFC| ≥1.0. Finally, the intersection was acquired from the Venn diagram online drawing tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
GO and KEGG pathway analysis of DEGs
The GO analysis is a shared method for large-scale functional enrichment research, in which gene functions can be divided into cellular component (CC), MF, and BP. The KEGG is a common biological information database. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) tool was conducted to perform GO and KEGG pathway analysis on DEGs (https://david.ncifcrf.gov/). Statistical significance was considered when P<0.01 and the gene count was ≥10.
Construction of PPI network and selection of hub genes
The Search Tool for the Retrieval of Interacting Genes (STRING), an open online tool (http://string-db.org/), was employed to analyze PPI information. A combined score was >0.4. Then, the PPI network was constructed by Cytoscape software. Finally, the degree of each protein node was calculated using the cytohubba plugin of Cytoscape. The top 10 genes were defined as hub genes of the network in our study. Clustering of the top 10 genes was constructed using an open multi-omics and clinical/phenotypic data exploration tool, UCSC Xena (http://xena.ucsc.edu/) (10).
Survival analysis of hub genes
The mRNA lung cancer database was used to evaluate the value of hub genes on survival and prognosis of LUAD patients by Kaplan-Meier plotter (http://kmplot.com/analysis/). The Kaplan-Meier plotter is an open online tool that mainly analyzes the impact of 54,675 genes on the survival and prognosis of patients in most tumor types. The value of hub genes for survival and prognosis in LUAD patients can be assessed using the mRNA lung cancer database in Kaplan-Meier plotter. The gene probe was based on “only jetset best probe set”. According to the median of mRNA expression, tumor patients were divided into high expression group and low expression group. Statistical significance was considered when P<0.01. The immunohistochemical data of 9 hub genes in LUAD or normal lung tissue were obtained from the Human Protein Atlas (HPA) (https://www.proteinatlas.org/).
PrognoScan database analysis
PrognoScan database is an online database (http://dna00.bio.kyutech.ac.jp/PrognoScan/), which uses a large number of cancer microarray dataset for the analysis of gene expression and relations between survival prognosis of patients. FGF2 expression and survival in patients with LUAD were analyzed in the PrognoScan database. Cox P<0.05 was statistically significant.
TIMER database analysis
TIMER is a comprehensive online web tool for systematic analysis of immune infiltration in different cancer types (https://cistrome.shinyapps.io/timer/). The DiffExp tool in TIMER was used to study the difference of FGF2 expression between tumor and normal tissues in The Cancer Genome Atlas (TCGA) tumor database. The gene module visualized the correlation between FGF2 expression and immune infiltration level. The results included the correlation analysis with B cells, CD4+ T cells, CD8+ T cells, Neutrophils, macrophages, and dendritic cells. The scatter plots were used to show the correlation between FGF2 and different immune cell infiltration. The gene expression level was determined by log2 RSEM.
Statistical analysis
The GEO2R is used to screen DEGs. The functional enrichment research of DEGs uses GO analysis. The STRING and Cytoscape were used to construct PPI network. The Kaplan-Meier Plotter database and Prognoscan database were used to analyze the survival of hub gene. The correlation between gene expression and immune infiltration was displayed by Spearman correlation and statistical significance. The correlation of variables is established by the following absolute values: 0.00–0.19 “very weak”, 0.20–0.39 “weak”, 0.40–0.59 “medium”. P<0.05 was statistically significant.
Results
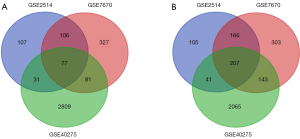
The DEGs were retained in LUAD. Through the analysis of GSE2514, GSE7670, and GSE40275, we obtained 840, 1,410, and 5,454 DEGs, respectively. Afterwards, the intersection of the DEGs of each dataset was taken by the Venn diagram, and 284 DEGs were obtained, of which 77 were highly expressed in tumor tissues and 207 were lowly expressed (Figure 1).
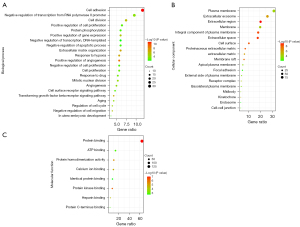
Functional enrichment of DEGs was carried out via GO and KEGG analysis. The GO analysis revealed that DEGs were mainly enriched in BPs such as cell adhesion, cell division, extracellular matrix organization, positive regulation of gene expression, positive regulation of angiogenesis, response to hypoxia, and so on. CC was mainly enriched in extracellular exosome, extracellular region, plasma membrane, membrane, integral component of plasma membrane, extracellular space, and so on. MF was primarily involved protein binding, protein kinase binding, protein homodimerization activity, calcium ion binding, and heparin binding execution. The details are shown in Table 1 and Figure 2. The KEGG analysis was mainly related to the cell cycle (Table 1).
Table 1
| Category | Term | Description | Count | P value |
|---|---|---|---|---|
| BP Term | GO:0007155 | Cell adhesion | 33 | 7.79e−12 |
| BP Term | GO:0051301 | Cell division | 21 | 1.83e−06 |
| BP Term | GO:0030198 | Extracellular matrix organization | 17 | 1.92e−07 |
| BP Term | GO:0010628 | Positive regulation of gene expression | 17 | 8.76e−06 |
| BP Term | GO:0045766 | Positive regulation of angiogenesis | 16 | 7.94e−10 |
| BP Term | GO:0001666 | Response to hypoxia | 16 | 1.97e−07 |
| BP Term | GO:0007179 | Transforming growth factor beta receptor signaling pathway | 12 | 3.70e−07 |
| BP Term | GO:0030336 | Negative regulation of cell migration | 11 | 4.11e−06 |
| BP Term | GO:0051726 | Regulation of cell cycle | 11 | 4.34e−05 |
| CC Term | GO:0005886 | Plasma membrane | 92 | 9.65e−05 |
| CC Term | GO:0070062 | Extracellular exosome | 76 | 7.16e−07 |
| CC Term | GO:0005576 | Extracellular region | 59 | 7.18e−10 |
| CC Term | GO:0016020 | Membrane | 58 | 5.25e−05 |
| CC Term | GO:0005887 | Integral component of plasma membrane | 53 | 3.36e−09 |
| CC Term | GO:0005615 | Extracellular space | 52 | 1.78e−09 |
| CC Term | GO:0009986 | Cell surface | 30 | 6.20e−09 |
| CC Term | GO:0005578 | Proteinaceous extracellular matrix | 21 | 6.95e−09 |
| CC Term | GO:0031012 | Extracellular matrix | 19 | 8.52e−07 |
| CC Term | GO:0045121 | Membrane raft | 18 | 2.21e−08 |
| CC Term | GO:0016324 | Apical plasma membrane | 15 | 1.80e−04 |
| CC Term | GO:0009897 | External side of plasma membrane | 12 | 4.88e−04 |
| CC Term | GO:0043235 | Receptor complex | 11 | 2.77e−05 |
| CC Term | GO:0016323 | Basolateral plasma membrane | 11 | 4.96e−04 |
| CC Term | GO:0000776 | Kinetochore | 10 | 4.29e−06 |
| CC Term | GO:0030496 | Midbody | 10 | 1.77e−04 |
| MF Term | GO:0005515 | Protein binding | 181 | 9.00e−08 |
| MF Term | GO:0019901 | Protein kinase binding | 22 | 5.44e−07 |
| MF Term | GO:0042803 | Protein homodimerization activity | 30 | 4.36e−06 |
| MF Term | GO:0005509 | Calcium ion binding | 28 | 2.46e−05 |
| MF Term | GO:0008201 | Heparin binding | 12 | 4.57e−05 |
| KEGG pathway | hsa04110 | Cell cycle | 11 | 6.77e−04 |
GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; LUAD, lung adenocarcinoma; BP, biological process; CC, cellular component; MF, molecular function.
The PPI network was constructed and hub genes were extracted. The PPI network consisted of 262 nodes and 1,363 interaction lines, in which the red nodes were highly expressed genes, and the green nodes were lowly expressed genes (Figure 3).
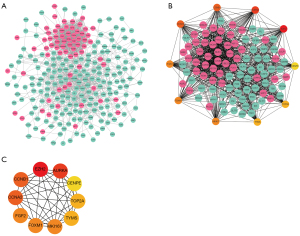
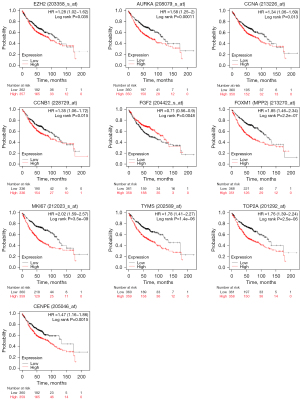
Next, the PPI networks with connectivity degree were further screened out. The top 10 genes were obtained, namely enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), aurora kinase A (AURKA), cyclin A2 (CCNA2), cyclin B1 (CCNB1), FGF2, forkhead box M1 (FOXM1), marker of proliferation Ki-67 (MKI67), thymidylate synthetase (TYMS), topoisomerase II alpha (TOP2A), and centromere protein E (CENPE) (Table 2), and a network diagram of their interaction was drawn (Figure 3). The results of hierarchical clustering showed that the hub genes could basically distinguish LUAD from normal lung tissue (Figure 4).
Table 2
| Gene symbol | Degree | Gene description |
|---|---|---|
| EZH2 | 43 | Enhancer of zeste 2 polycomb repressive complex 2 subunit |
| AURKA | 41 | Aurora kinase A |
| CCNA2 | 40 | Cyclin A2 |
| CCNB1 | 40 | Cyclin B1 |
| FGF2 | 39 | Fibroblast growth factor 2 |
| FOXM1 | 39 | Forkhead box M1 |
| MKI67 | 39 | Marker of proliferation Ki-67 |
| TYMS | 38 | Thymidylate synthetase |
| TOP2A | 38 | Topoisomerase (DNA) II alpha |
| CENPE | 37 | Centromere protein E |
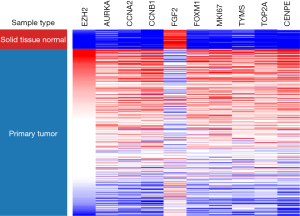
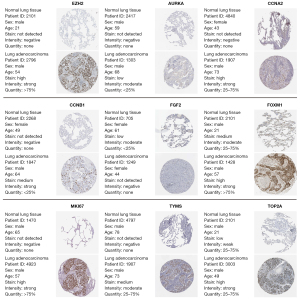
Survival analyses of hub genes were performed. According to the hub genes expression, the survival curves of LUAD patients were drawn via Kaplan-Meier plotter (Figure 5). It was found that patients with high expression of EZH2, AURKA, CCNA2, CCNB1, FOXM1, MKI67, TYMS, TOP2A, and GEPNE have a poor prognosis, while patients with high FGF2 gene expression have a better prognosis, which is consistent with the previous expression results in the mRNA datasets. The expression of hub genes in LUAD and normal lung tissues was obtained from immunohistochemical data of HPA (Figure 6). The expression results of the hub genes in different sample types of immunohistochemistry were consistent with the results of the above database.
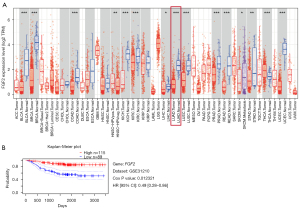
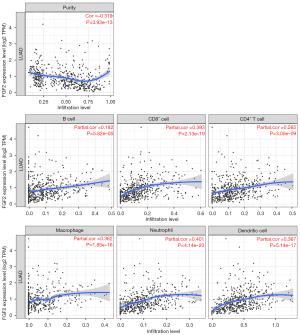
FGF2 and tumor immune microenvironment. We analyzed the expression of FGF2 in TCGA database by TIMER web service. It also found that FGF2 was highly expressed in normal tissues compared with tumor tissues (Figure 7A). We further confirmed that the high expression of FGF2 was associated with better prognosis in LUAD patients in PrognoScan database (Figure 7B). Then, TIMER was used to analyze the relationship between FGF2 and tumor purity in LUAD, and it was found that the expression level of FGF2 was negatively correlated with tumor purity (Figure 8). The expression level of FGF2 was found to be positively correlated with the immune infiltration of B cells, CD4+ T cells, CD8+ T cells, Neutrophils, macrophages, and dendritic cells. There was moderate correlation with neutrophils infiltration, but weak correlation with CD4+ T cells, CD8+ T cells, macrophages, and dendritic cells infiltration.
Discussion
The most common type of lung cancer is NSCLC, accounting for 85% of all lung cancer cases. As the most common subtype of NSCLC, LUAD comprises approximately 40% of the cases (11), and the incidence of LUAD is increasing annually. Although there has been significant improvement in the diagnosis and treatment strategies for LUAD, such as targeted treatments, the long-term survival rate of patients has remained low. The main reason is the dynamic drug resistance of the tumors (12). Therefore, it is urgent that the pathogenesis of LUAD be further studied in order to find new diagnostic and therapeutic targets. With the development of bioinformatics, the screening of DEGs provides a valuable tool for research. We may find new gene targets by comparing and extracting a large-scale LUAD mRNA expression dataset.
In this study, we screened 284 DEGs by comparing 3 datasets, including 77 high expression genes and 207 low expression genes. After GO analysis of 284 DEGs, it was found that their biological functions were mainly concerned cell adhesion and division, while KEGG pathway was enriched in the cell cycle. We further extracted the top 10 hub genes by PPI network, which were EZH2, AURKA, CCNA2, CCNB1, FGF2, FOXM1, MKI67, TYMS, TOP2A, and CENPE. Among them, FGF2 was a low-expression gene, and the others were high expression genes. Finally, survival analysis showed that the expression results of hub genes were consistent in patients, and the prognosis was strongly correlated with the hub gene expression.
The EZH2 gene is a component of polycomb repressive complex 2 (PRC2), a methyltransferase. Increasing DNA methylation and inactivating tumor suppressor genes are important factors by which EZH2 promotes tumor progression (13,14). It can also enhance cell proliferation by promoting cell cycle processes (15). A study found that EZH2 acetylation enhanced the metastasis and invasion ability of tumor cells and that it was correlated with poor prognoses in LUAD patients (16). It has been reported that the vascular endothelial growth factor (VEGF)/vascular endothelial growth factor receptor 2 (VEGFR-2) pathway can regulate the EZH2 expression in LUAD (17).
The AURKA gene is a serine/threonine kinase that plays a key role in regulating the cell cycle and mitosis of normal cells (18), and its abnormal expression has been also correlated with various types of tumors (19-21). Some studies have found that the activation of AURKA in NSCLC can increase the resistance to anti-EGFR targeted therapies (22,23). In addition, the differential expression of AURKA in LUAD has been confirmed by previous studies (24,25). In our study, we also found that AURKA was highly expressed in adenocarcinoma, and the high expression of AURKA in LUAD patients was obviously correlated with poor prognosis. Therefore, the level of AURKA expression may be used as a diagnosis tool and treatment target for LUAD. The expression of AURKA can be further studied for its role in the development and progression of LUAD.
The FGF2 gene is a member of the fibroblast growth factor family. It mainly binds FGFR on the cell surface and is involved in cell proliferation, metastasis, and angiogenesis. In previous studies, it was found that FGF2 expression in NSCLC was negatively correlated with the survival rate (26,27). However, in our study, we found that FGF2 was lowly expressed in LUAD tissues, and survival analysis also showed that patients with low FGF2 expression had better prognoses. This shows that the mechanism of FGF2 in LUAD is not fully clarified, and further study should be undertaken. Therefore, we further investigated the mechanism of FGF2 affects the prognosis of LUAD patients. It is known that the immune microenvironment plays an important role in NSCLC, and immunotherapy (PD-L1 inhibitors) has a significant effect in the treatment of lung tumors (28,29). So, we analyzed the effect of FGF2 expression on the immune microenvironment in LUAD. It is noteworthy that this study found that the high FGF2 expression decreased tumor purity, and the FGF2 expression level was positively correlated with immune cell infiltration. Although the correlation was not very strong, the significance was very high (Figure 8). Thus, we can appropriately believe that immune infiltration is one of the reasons for the better prognosis of patients with high FGF2 expression.
The FOXM1 gene is a transcription factor of the forkhead box family. Its high expression is associated with various types of tumors. It plays an important role in cell proliferation, metastasis, drug resistance, and the promotion of vascular survival and cell cycling (30-32). In addition, studies have found that FOXM1 is correlated with the development of LUAD, being specifically associated with its proliferation, invasion, and metastasis (33-35).
The CENPE gene is a kinesin-like motor protein, and its overexpression is related to the development and progression of tumors (36). In NSCLC, the high expression of CENPE was correlated with the negative prognoses of patients (37). In addition, studies have found that CENPE can directly act on FOXM1 to regulate the proliferation of LUAD cells (33).
The TOP2A gene mainly acts on DNA transcription and replication enzymes, and its overexpression is associated with many tumors. Also, studies have shown that TOP2A can target CCNB1 and CCNB2 in LUAD to promote tumor proliferation and metastasis (38), leading to poor patient prognosis (39). In addition, CCNA2 and CCNB1 are proteins that can regulate the cell cycle. Their abnormal expression was correlated with the development and progression of NSCLC and poor patient prognosis (40-42). For example, CCNA2 can promote the epithelial-mesenchymal transition of NSCLC cells via integrin (43).
The TYMS gene is a key enzyme that maintains DNA synthesis and repair, and is considered one of the targets of new antifolate drugs such as pemetrexed (44). Its high expression was also linked to the poor prognosis of NSCLC (45).
The expression of MKI67 is related to the proliferation and growth of tumors. It is widely used as a proliferation and prognostic marker for research (46). It is also highly expressed in lung cancer (46-48).
In summary, 284 DEGs and 10 hub genes were obtained from the GEO database in our study. These 10 hub genes were strongly correlated with patient prognosis. Previous studies have demonstrated that the abnormal expression of hub genes was often strongly correlated with the development and prognosis of lung cancer, which is consistent with our results with LUAD. In the future, genes with high expression of EZH2, AURKA, CCNA2, CCNB1, FGF2, FOXM1, MKI67, TYMS, TOP2A, and CENPE may be used as targets for tumor therapy to prolong the survival time of LUAD patients. The relationship between FGF2 and immune microenvironment can also be explored to provide new strategies for the study and treatment of LUAD. However, some detailed mechanisms of hub genes in LUAD remain unclear and require further study in the future. In summary, we screened the GEO database to obtain hub genes that were highly correlated with LUAD. These genes may be potential biomarkers or therapeutic targets for LUAD.
We used some bioinformatics methods to find key genes via this study. However, due to the technical condition limitations, some other bioinformatics hot issues include weighted gene co-expression network analysis, competing endogenous RNA (ceRNA) network, DNA repair gene mutations and increased Neoantigen load and activated T cell infiltration were not carried out in this study, which will be the direction of our future research in LUAD. At the same time, as a result of the limitation of basic experimental conditions, this research was limited to the analysis of open database, and did not conduct functional research on hub genes. It is necessary to explore the mechanism of hub genes in the development of LUAD through functional research in the future.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2676/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2676/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- She J, Yang P, Hong Q, et al. Lung cancer in China: challenges and interventions. Chest 2013;143:1117-26. [Crossref] [PubMed]
- Wang LQ, Zhao LH, Qiao YZ. Identification of potential therapeutic targets for lung cancer by bioinformatics analysis. Mol Med Rep 2016;13:1975-82. [Crossref] [PubMed]
- Mitchell KA, Zingone A, Toulabi L, et al. Comparative Transcriptome Profiling Reveals Coding and Noncoding RNA Differences in NSCLC from African Americans and European Americans. Clin Cancer Res 2017;23:7412-25. [Crossref] [PubMed]
- Rades D, Setter C, Dahl O, et al. Fibroblast growth factor 2--a predictor of outcome for patients irradiated for stage II-III non-small-cell lung cancer. Int J Radiat Oncol Biol Phys 2012;82:442-7. [Crossref] [PubMed]
- Hegab AE, Ozaki M, Kameyama N, et al. Effect of FGF/FGFR pathway blocking on lung adenocarcinoma and its cancer-associated fibroblasts. J Pathol 2019;249:193-205. [Crossref] [PubMed]
- Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol 2005;167:1763-75. [Crossref] [PubMed]
- Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics 2007;8:140. [Crossref] [PubMed]
- Kastner S, Voss T, Keuerleber S, et al. Expression of G protein-coupled receptor 19 in human lung cancer cells is triggered by entry into S-phase and supports G(2)-M cell-cycle progression. Mol Cancer Res 2012;10:1343-58. [Crossref] [PubMed]
- Goldman MJ, Craft B, Hastie M, et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 2020;38:675-8. [Crossref] [PubMed]
- Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. [Crossref] [PubMed]
- Callegari D, Ranaghan KE, Woods CJ, et al. L718Q mutant EGFR escapes covalent inhibition by stabilizing a non-reactive conformation of the lung cancer drug osimertinib. Chem Sci 2018;9:2740-9. [Crossref] [PubMed]
- Cooper WA, Lam DC, O'Toole SA, et al. Molecular biology of lung cancer. J Thorac Dis 2013;5:S479-90. [PubMed]
- Jakopovic M, Thomas A, Balasubramaniam S, et al. Targeting the epigenome in lung cancer: expanding approaches to epigenetic therapy. Front Oncol 2013;3:261. [Crossref] [PubMed]
- Xia H, Zhang W, Li Y, et al. EZH2 silencing with RNA interference induces G2/M arrest in human lung cancer cells in vitro. Biomed Res Int 2014;2014:348728. [Crossref] [PubMed]
- Wan J, Zhan J, Li S, et al. PCAF-primed EZH2 acetylation regulates its stability and promotes lung adenocarcinoma progression. Nucleic Acids Res 2015;43:3591-604. [Crossref] [PubMed]
- Riquelme E, Suraokar M, Behrens C, et al. VEGF/VEGFR-2 upregulates EZH2 expression in lung adenocarcinoma cells and EZH2 depletion enhances the response to platinum-based and VEGFR-2-targeted therapy. Clin Cancer Res 2014;20:3849-61. [Crossref] [PubMed]
- Nishimura Y, Endo T, Kano K, et al. Porcine Aurora A accelerates Cyclin B and Mos synthesis and promotes meiotic resumption of porcine oocytes. Anim Reprod Sci 2009;113:114-24. [Crossref] [PubMed]
- Katsha A, Belkhiri A, Goff L, et al. Aurora kinase A in gastrointestinal cancers: time to target. Mol Cancer 2015;14:106. [Crossref] [PubMed]
- Bischoff JR, Anderson L, Zhu Y, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J 1998;17:3052-65. [Crossref] [PubMed]
- Donnella HJ, Webber JT, Levin RS, et al. Kinome rewiring reveals AURKA limits PI3K-pathway inhibitor efficacy in breast cancer. Nat Chem Biol 2018;14:768-77. [Crossref] [PubMed]
- Bertran-Alamillo J, Cattan V, Schoumacher M, et al. AURKB as a target in non-small cell lung cancer with acquired resistance to anti-EGFR therapy. Nat Commun 2019;10:1812. [Crossref] [PubMed]
- Shah KN, Bhatt R, Rotow J, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med 2019;25:111-8. [Crossref] [PubMed]
- Zhang MY, Liu XX, Li H, et al. Elevated mRNA Levels of AURKA, CDC20 and TPX2 are associated with poor prognosis of smoking related lung adenocarcinoma using bioinformatics analysis. Int J Med Sci 2018;15:1676-85. [Crossref] [PubMed]
- Selvaraj G, Kaliamurthi S, Kaushik AC, et al. Identification of target gene and prognostic evaluation for lung adenocarcinoma using gene expression meta-analysis, network analysis and neural network algorithms. J Biomed Inform 2018;86:120-34. [Crossref] [PubMed]
- Iwasaki A, Kuwahara M, Yoshinaga Y, et al. Basic fibroblast growth factor (bFGF) and vascular endothelial growth factor (VEGF) levels, as prognostic indicators in NSCLC. Eur J Cardiothorac Surg 2004;25:443-8. [Crossref] [PubMed]
- Donnem T, Al-Shibli K, Al-Saad S, et al. Prognostic impact of fibroblast growth factor 2 in non-small cell lung cancer: coexpression with VEGFR-3 and PDGF-B predicts poor survival. J Thorac Oncol 2009;4:578-85. [Crossref] [PubMed]
- Barbee MS, Ogunniyi A, Horvat TZ, et al. Current status and future directions of the immune checkpoint inhibitors ipilimumab, pembrolizumab, and nivolumab in oncology. Ann Pharmacother 2015;49:907-37. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Zhang Y, Zhang N, Dai B, et al. FoxM1B transcriptionally regulates vascular endothelial growth factor expression and promotes the angiogenesis and growth of glioma cells. Cancer Res 2008;68:8733-42. [Crossref] [PubMed]
- Li Q, Zhang N, Jia Z, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res 2009;69:3501-9. [Crossref] [PubMed]
- Monteiro LJ, Khongkow P, Kongsema M, et al. The Forkhead Box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene 2013;32:4634-45. [Crossref] [PubMed]
- Shan L, Zhao M, Lu Y, et al. CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. Int J Oncol 2019;55:257-66. [PubMed]
- Wei P, Zhang N, Wang Y, et al. FOXM1 promotes lung adenocarcinoma invasion and metastasis by upregulating SNAIL. Int J Biol Sci 2015;11:186-98. [Crossref] [PubMed]
- Chao Y, Shang J, Ji W. ALKBH5-m6A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Biophys Res Commun 2020;521:499-506. [Crossref] [PubMed]
- Hitti E, Bakheet T, Al-Souhibani N, et al. Systematic Analysis of AU-Rich Element Expression in Cancer Reveals Common Functional Clusters Regulated by Key RNA-Binding Proteins. Cancer Res 2016;76:4068-80. [Crossref] [PubMed]
- Hao X, Qu T. Expression of CENPE and its Prognostic Role in Non-small Cell Lung Cancer. Open Med (Wars) 2019;14:497-502. [Crossref] [PubMed]
- Kou F, Sun H, Wu L, et al. TOP2A Promotes Lung Adenocarcinoma Cells' Malignant Progression and Predicts Poor Prognosis in Lung Adenocarcinoma. J Cancer 2020;11:2496-508. [Crossref] [PubMed]
- Guo W, Sun S, Guo L, et al. Elevated TOP2A and UBE2C expressions correlate with poor prognosis in patients with surgically resected lung adenocarcinoma: a study based on immunohistochemical analysis and bioinformatics. J Cancer Res Clin Oncol 2020;146:821-41. [Crossref] [PubMed]
- Ko E, Kim Y, Cho EY, et al. Synergistic effect of Bcl-2 and cyclin A2 on adverse recurrence-free survival in stage I non-small cell lung cancer. Ann Surg Oncol 2013;20:1005-12. [Crossref] [PubMed]
- Cooper WA, Kohonen-Corish MR, McCaughan B, et al. Expression and prognostic significance of cyclin B1 and cyclin A in non-small cell lung cancer. Histopathology 2009;55:28-36. [Crossref] [PubMed]
- Yoshida T, Tanaka S, Mogi A, et al. The clinical significance of Cyclin B1 and Wee1 expression in non-small-cell lung cancer. Ann Oncol 2004;15:252-6. [Crossref] [PubMed]
- Ruan JS, Zhou H, Yang L, et al. CCNA2 facilitates epithelial-to-mesenchymal transition via the integrin αvβ3 signaling in NSCLC. Int J Clin Exp Pathol 2017;10:8324-33. [PubMed]
- Chen CY, Chang YL, Shih JY, et al. Thymidylate synthase and dihydrofolate reductase expression in non-small cell lung carcinoma: the association with treatment efficacy of pemetrexed. Lung Cancer 2011;74:132-8. [Crossref] [PubMed]
- Ceppi P, Rapa I, Lo Iacono M, et al. Expression and pharmacological inhibition of thymidylate synthase and Src kinase in nonsmall cell lung cancer. Int J Cancer 2012;130:1777-86. [Crossref] [PubMed]
- Folescu R, Levai CM, Grigoraş ML, et al. Expression and significance of Ki-67 in lung cancer. Rom J Morphol Embryol 2018;59:227-33. [PubMed]
- Ahn HK, Jung M, Ha SY, et al. Clinical significance of Ki-67 and p53 expression in curatively resected non-small cell lung cancer. Tumour Biol 2014;35:5735-40. [Crossref] [PubMed]
- Maddau C, Confortini M, Bisanzi S, et al. Prognostic significance of p53 and Ki-67 antigen expression in surgically treated non-small cell lung cancer: immunocytochemical detection with imprint cytology. Am J Clin Pathol 2006;125:425-31. [Crossref] [PubMed]
(English Language Editor: J. Jones)


