
Identification of genes and pathways leading to poor prognosis of non-small cell lung cancer using integrated bioinformatics analysis
Introduction
Lung cancer is one of the most common malignancies in the world (1). Lung cancer is classified as non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is significantly more common than SCLC and is further subdivided based on histology (e.g., adenocarcinoma, squamous or large cell carcinoma) (2,3). Despite the introduction of several new and more effective biomarkers in clinical practice, only 25% of NSCLC patients are diagnosed at stage I–II when NSCLC patients are still amenable to radical surgery (4). The 5-year survival is 77–92% for clinical stage IA, 68% for stage IB, 60% for stage IIA, and 53% for stage IIB (5). Radical surgery indeed improved the 5-year survival rates for patients with stage I–II NSCLC. However, most NSCLC patients are diagnosed at a late stage due to the typically asymptomatic early stage and the lack of effective screening trial. These patients with advanced NSCLC are not amenable to radical surgery, but up to 69% of them could have a potentially actionable molecular target (6). Epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) are the most common oncogenic drivers in NSCLC. Although EGFR tyrosine kinase inhibitor (TKI) and ALK TKI treatment show an encouraging improvement in overall survival, nearly all patients eventually have disease progression due to acquired resistance after treatment (7). The reduction of NSCLC mortality has been set as a major priority worldwide by detecting new molecular targets and prompting the development of new therapies.
Traditional gene-by-gene approaches in research are insufficient to meet the growth and demand of biological research in understanding true biology. In recent years, bioinformatics analysis has identified a group of cancer related genes that provide insight into the molecular mechanism of diseases progression (8,9). Nevertheless, there are few studies of NSCLC via these bioinformatics analyses. In this paper, bioinformatics analysis was utilized to explore the potential biomarkers and the molecular mechanism of NSCLC.
We present the following article in accordance with the REMARK reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1986/rc).
Methods
In this paper, original microarray datasets were downloaded from GEO. DEGs between NSCLC and adjacent lung tissue were filtered via the GEO2R online tool. The functions and pathway enrichment of the differentially expressed genes (DEGs) were identified via the DAVID online database. The protein-protein interactions (PPIs) of these DEGs were constructed by the STRING website and visualized by the Cytoscape software platform. The GEPIA database was used to evaluate the expression of genes. Kaplan-Meier plotter was used to analyse the survival rate with multivariate Cox regression. The expression of protein tyrosine kinase 2 (PTK2) was analysed via the UALCAN database. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Microarray data information and DEGs identification
NCBI-GEO (https://www.ncbi.nlm.nih.gov/gds/) is a functional genomics database that includes microarray and cutting edge sequencing. Four original microarray datasets (GSE33532, GSE19188, GSE102287, and GSE27262) were downloaded from the NCBI-GEO database, from which data on 319 NSCLC and 232 adjacent lung tissues was available. DEGs between NSCLC and adjacent lung tissue were filtered via the GEO2R online tool with P<0.05 and (logFC) >2 (10). The overlapping DEGs among the four datasets were identified through the Venn diagram database (http://bioinformatics.psb.ugent.be/webtools/Venn/). The DEGs with logFC >0 were the upregulated genes; in contrast, which gene was the downregulated gene with logFC <0.
Gene functional and pathways enrichment analysis
Gene ontology (GO), which comprises 3 independent ontologies (cellular component, molecular function, and biological process), is the most comprehensive and widely used knowledge base concerning the functions of genes (11). Kyoto Encyclopedia of Genes and Genomes (KEGG) is a database resource for understanding the high-level functions and utilities of biological systems (12). DAVID (https://david.ncifcrf.gov/home.jsp) is a comprehensive functional annotation tool for investigators to understand the biological meaning behind a large list of genes (13). In the present study, the functions and pathway enrichment of the DEGs were identified via the DAVID online database. P<0.05 was used as the cut-off criterion.
PPI network and module analysis
The PPI network of the proteins encoded by DEGs was constructed via the STRING database (https://cn.string-db.org/) (14). The Cytoscape software platform (https://cytoscape.org/) was utilized to construct and visualize the PPI network with a maximum number of inter actors =0 and confidence score ≥0.4 (15). The nodes represent the genes, and the edges between the nodes represent the interactions between the genes in the PPI network.
Expression and survival analysis of hub genes in NSCLC
The expression of hub genes between NSCLC and adjacent lung tissues was compared through the GEPIA database (http://gepia.cancer-pku.cn/) (16). NSCLC patients were divided into low- and high-expression groups according to the median expression of each hub gene. Kaplan-Meier analysis was performed via Kaplan-Meier plotter (https://kmplot.com/analysis/) (17). P value includes correction for multiple hypothesis testing. P<0.05 indicated that the difference was statistically significant.
Expression and survival analysis of PTK2 in NSCLC
The UALCAN database (http://ualcan.path.uab.edu/) was employed to analyse the expression of PTK2 between NSCLC and adjacent lung tissues (18). The survival prognosis of PTK2 was evaluated by Kaplan-Meier plotter.
Statistical analysis
Bioinformatics analyses of databases were described in detail in the above method. The two-sided Student’s t-test was used to compare the differences between groups. Differences between groups were compared using a two-sided Student’s t-test. Kaplan-Meier analysis was performed via Kaplan-Meier plotter. P<0.05 was considered statistically significant.
Results
Microarray data information and DEGs identification
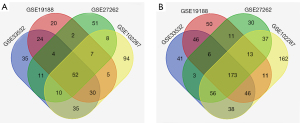
Four original microarray datasets (GSE33532, GSE19188, GSE102287 and GSE27262) were obtained from the NCBI-GEO database. A total of 3188 DEGs were extracted via the GEO2R online tool using P<0.05 and (logFC) >2 as cut-off criteria. A total of 225 common DEGs were identified through the Venn diagram database, including 52 upregulated genes and 173 downregulated genes in NSCLC tissue compared to adjacent lung tissues (Table 1, Figure 1).
Table 1
| DEG | Gene symbol |
|---|---|
| Upregulated | ADAM12, TPX2, CCNB1, SULF1, HMGB3, ASPM, FERMT1, HMMR, CXCL13, KIF4A, GINS1, TMPRSS4, HS6ST2, SPP1, COL1A1, ADAMDEC1, ANLN, BIRC5, KIF20A, UBE2C, COL10A1, CCNB2, PSAT1, TYMS, CDCA7, MELK, COL11A1, KIF11, CEP55, CDC20, CTHRC1, RRM2, ZWINT, TOP2A, KIAA0101, GJB2, GREM1, TTK, GTSE1, CDKN3, BUB1, NUF2, CENPU, MMP1, NEK2, MMP12, AURKA, UBE2T, CENPF, TFAP2A, MAD2L1, DLGAP5 |
| Downregulated | HBA2///HBA1, EDN1, RTKN2, EMCN, SOX7, ADARB1, CHRDL1, PPP1R14A, ADGRD1, GPIHBP1, KCNT2, MFAP4, PEBP4, ITIH5, ERG, SLC6A4, PECAM1, KCNK3, MMRN2, NOSTRIN, SYNPO2, NCKAP5, GIMAP8, OGN, SCARA5, BTNL9, PCAT19, IGSF10, ACVRL1, SCGB1A1, CDO1, CA4, SDPR, WWC2///CLDN22, TEK, CLIC3, GRK5, ID4, EXOSC7///CLEC3B, PLA2G1B, DACH1, VGLL3, FAM150B, ANOS1, ACKR1, LIFR, STXBP6, S1PR1, EMP2, LYVE1, ADAMTS8, HBEGF, PTPN21, GDF10, LAMP3, LIMCH1, LEPROT///LEPR, DNASE1L3, BCHE, SPOCK2, AKAP12, CD36, FAM162B, PDE5A, LDB2, ROBO4, SPTBN1, CALCRL, CAV1, TBX5-AS1, PPBP, JAM2, PTPRB, QKI, FOXF1, ACADL, ANKRD29, PIR-FIGF///FIGF, AQP4, NEBL, ITGA8, MT1M, TNNC1, PDZD2, FAT3, ADIRF, MCEMP1, HBB, FHL1, RHOJ, CPB2, SRPX, FAM189A2, SORBS2, LRRN3, THBD, KLF4, EMP1, FMO2, ABCA8, MYZAP, SOCS2, SLC39A8, AOC3, SFTPC, ADRB1, SEMA3G, TCF21, NEDD4L, TGFBR3, HHIP, PGC, ADH1B, ARHGEF26, ARHGAP6, LPL, ASPA, FABP4, EDNRB, SOSTDC1, SCN4B, FCN3, MYCT1, KANK3, DLC1, STX11, LINC00312, FAM107A, CCDC85A, PLAC9, CCBE1, PGM5, C1QTNF7, GPX3, AGER, FOSB, RGCC, VWF, SEMA5A, PIP5K1B, ABI3BP, CD93, BMP2, TIE1, KIAA1462, VIPR1, AGTR1, WIF1, EPAS1, RAMP3, CLIC5, NPNT, SLIT2, GIMAP6, FHL5, MAMDC2, ADAMTSL3, CLDN18, C2orf40, CDH5, PDK4, GPM6A, COL6A6, FILIP1, CFD, GKN2, ANGPT1, CYP4B1, SMAD6, HYAL1, TMEM100, DUOX1, AFF3 |
DEG, differentially expressed gene.
Gene function and pathways enrichment analysis
Functions and pathway enrichment of DEGs were conducted using the DAVID database. The DEGs were classified into three functional groups: biological process group (BP), cellular component group (CC) and molecular function group (MF) (Figure 2, Table 2). In the BP group, the cell division GO term enriched both upregulated and downregulated DEGs, which indicates that the biological process of cell division may play vital roles in the development of NSCLC. In addition, the upregulated DEGs were also involved in mitotic nuclear division and sister chromatid cohesion, while the downregulated DEGs were involved in angiogenesis and the BMP signaling pathway. In the CC group, the upregulated DEGs were enriched in spindle, midbody and condensed chromosome kinetochore, while the downregulated DEGs were enriched in cell surface, extracellular region and extracellular space. In the MF group, the overexpressed DEGs mainly included molecular functions of ATP binding, metalloendopeptidase activity and protein homodimerization activity, whereas the downregulated genes included those of heparin binding, receptor activity and transforming growth factor beta binding.
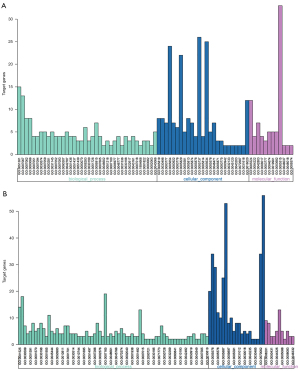
Table 2
| Expression | Category | Term | Count | % | P value | FDR |
|---|---|---|---|---|---|---|
| Upregulated | GOTERM_BP_DIRECT | GO:0051301 | 15 | 17.04 | 1.45E-12 | 2.07E-09 |
| ~cell division | ||||||
| GOTERM_BP_DIRECT | GO:0007067 | 13 | 14.77 | 7.87E-12 | 1.12E-08 | |
| ~mitotic nuclear division | ||||||
| GOTERM_BP_DIRECT | GO:0007062 | 8 | 9.09 | 2.47E-08 | 3.52E-05 | |
| ~sister chromatid cohesion | ||||||
| GOTERM_BP_DIRECT | GO:0000086 | 8 | 9.09 | 1.77E-07 | 2.52E-04 | |
| ~G2/M transition of mitotic cell cycle | ||||||
| GOTERM_BP_DIRECT | GO:0007051 | 4 | 4.54 | 1.44E-05 | 0.02049 | |
| ~spindle organization | ||||||
| GOTERM_BP_DIRECT | GO:0007094 | 4 | 4.54 | 2.90E-05 | 0.04136 | |
| ~mitotic spindle assembly checkpoint | ||||||
| GOTERM_CC_DIRECT | GO:0005819 | 8 | 9.09 | 4.34E-08 | 4.77E-05 | |
| ~spindle | ||||||
| GOTERM_CC_DIRECT | GO:0030496 | 8 | 9.09 | 6.76E-08 | 7.43E-05 | |
| ~midbody | ||||||
| GOTERM_CC_DIRECT | GO:0000777 | 7 | 7.95 | 1.51E-07 | 1.66E-04 | |
| ~condensed chromosome kinetochore | ||||||
| GOTERM_CC_DIRECT | GO:0005654 | 24 | 27.27 | 3.90E-07 | 4.28E-04 | |
| ~nucleoplasm | ||||||
| GOTERM_CC_DIRECT | GO:0000922 | 7 | 7.95 | 5.77E-07 | 6.34E-04 | |
| ~spindle pole | ||||||
| GOTERM_CC_DIRECT | GO:0000776 | 6 | 6.82 | 3.06E-06 | 0.00337 | |
| ~kinetochore | ||||||
| GOTERM_MF_DIRECT | GO:0005524 | 12 | 13.63 | 0.00316 | 3.56706 | |
| ~ATP binding | ||||||
| GOTERM_MF_DIRECT | GO:0004222 | 4 | 4.54 | 0.00430 | 4.82813 | |
| ~metalloendopeptidase activity | ||||||
| GOTERM_MF_DIRECT | GO:0042803 | 7 | 7.95 | 0.01842 | 19.21940 | |
| ~protein homodimerization activity | ||||||
| Downregulated | GOTERM_BP_DIRECT | GO:0001525 | 14 | 5.37 | 8.86E-08 | 1.44E-04 |
| ~angiogenesis | ||||||
| GOTERM_BP_DIRECT | GO:0007155 | 18 | 6.91 | 6.24E-07 | 0.00101 | |
| ~cell adhesion | ||||||
| GOTERM_BP_DIRECT | GO:0030509 | 7 | 2.69 | 5.57E-05 | 0.09039 | |
| ~BMP signaling pathway | ||||||
| GOTERM_BP_DIRECT | GO:0042310 | 4 | 1.54 | 4.17E-04 | 0.67403 | |
| ~vasoconstriction | ||||||
| GOTERM_BP_DIRECT | GO:0051591 | 5 | 1.92 | 7.09E-04 | 1.14466 | |
| ~response to cAMP | ||||||
| GOTERM_BP_DIRECT | GO:0001666 | 8 | 3.07 | 8.34E-04 | 1.34494 | |
| ~response to hypoxia | ||||||
| GOTERM_CC_DIRECT | GO:0009986 | 20 | 7.68 | 2.72E-07 | 3.30E-04 | |
| ~cell surface | ||||||
| GOTERM_CC_DIRECT | GO:0005576 | 34 | 13.05 | 2.85E-06 | 0.00345 | |
| ~extracellular region | ||||||
| GOTERM_CC_DIRECT | GO:0005615 | 29 | 11.13 | 1.43E-05 | 0.01737 | |
| ~extracellular space | ||||||
| GOTERM_CC_DIRECT | GO:0005578 | 12 | 4.61 | 2.40E-05 | 0.02907 | |
| ~proteinaceous extracellular matrix | ||||||
| GOTERM_CC_DIRECT | GO:0045121 | 10 | 3.84 | 8.31E-05 | 0.10057 | |
| ~membrane raft | ||||||
| GOTERM_MF_DIRECT | GO:0008201 | 9 | 3.45 | 5.11E-05 | 0.06988 | |
| ~heparin binding | ||||||
| GOTERM_MF_DIRECT | GO:0004872 | 8 | 3.07 | 0.00206 | 2.77954 | |
| ~receptor activity | ||||||
| GOTERM_MF_DIRECT | GO:0050431 | 3 | 1.15 | 0.00739 | 9.63795 | |
| ~transforming growth factor beta binding |
GO, gene ontology; NSCLC, non-small cell lung cancer; FDR, false discovery rate; BMP, bone morphogenetic protein.
As shown in Table 3, KEGG analysis results demonstrated that the upregulated DEGs were significantly enriched in oocyte meiosis, the cell cycle and the p53 signaling pathway, while the downregulated DEGs were particularly enriched in malaria, vascular smooth muscle contraction and the PPAR signaling pathway.
Table 3
| Expression | Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|---|
| Upregulated | hsa04114 | Oocyte meiosis | 6 | 6.82 | 2.11E-05 | CCNB1, MAD2L1, CCNB2, BUB1, AURKA, CDC20 |
| hsa04110 | Cell cycle | 6 | 6.82 | 3.62E-05 | CCNB1, MAD2L1, CCNB2, BUB1, TTK, CDC20 | |
| hsa04115 | p53 signaling pathway | 4 | 4.55 | 0.00119 | CCNB1, CCNB2, RRM2, GTSE1 | |
| hsa04914 | Progesterone-mediated oocyte maturation | 4 | 4.55 | 0.00253 | CCNB1, MAD2L1, CCNB2, BUB1 | |
| hsa04512 | ECM-receptor interaction | 4 | 4.55 | 0.00253 | COL1A1, COL11A1, SPP1, HMMR | |
| hsa04974 | Protein digestion and absorption | 3 | 3.41 | 0.03166 | COL1A1, COL11A1, COL10A1 | |
| Downregulated | hsa05144 | Malaria | 4 | 1.54 | 0.01182 | CD36, PECAM1, ACKR1, HBB |
| hsa04270 | Vascular smooth muscle contraction | 5 | 1.92 | 0.02681 | RAMP3, AGTR1, PLA2G1B, CALCRL, PPP1R14A | |
| hsa03320 | PPAR signaling pathway | 4 | 1.54 | 0.02717 | LPL, CD36, FABP4, ACADL | |
| hsa04610 | Complement and coagulation cascades | 4 | 1.54 | 0.02931 | VWF, THBD, CFD, CPB2 | |
| hsa04514 | CAMs | 5 | 1.92 | 0.04910 | CLDN18, ITGA8, PECAM1, JAM2, CDH5 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; NSCLC, non-small cell lung cancer; ECM, extracellular matrix; PDAR, peroxisome proliferator-activated receptor; CAMs, cell adhesion molecules.
PPI network and module analysis
A total of 174 DEGs, including 48 upregulated genes and 126 downregulated genes, were filtered into the PPI network complex, which included 174 nodes and 816 edges, via the STRING database and Cytoscape software platform (Figure 3A). There were 51 of the 225 DEGs that failed to fall into the PPI network. Two significant modules in the PPI network complex were collected for further analysis using the MCODE plugin of the Cytotype software platform. A total of 32 central nodes and 511 edges were identified in module 1, and all 32 central nodes were upregulated genes and mainly associated with the oocyte meiosis, the cell cycle, the p53 signaling pathway and the progesterone-mediated oocyte maturation (Figure 3B, Table 4). In addition, module 2, which included 9 nodes and 29 edges, was mainly associated with the PI3K-Akt and HIF-1 signaling pathways (Figure 3C, Table 4). The CytoHubba plugin was used to identify the top 10 hub genes (COL1A1, ITGA8, VWF, MMP1, ADAM12, CD36, OGN, EDN1, CTHRC1 and CAV) in the PPI network (Figure 3D, Table 4), which were mainly involved in ECM-receptor interaction, focal adhesion, and the PI3K-Akt signaling pathway.
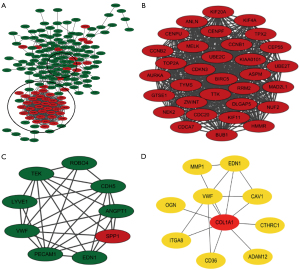
Table 4
| Module | Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|---|
| Module 1 | hsa04114 | Oocyte meiosis pathway | 6 | 10.43 | 1.16E-06 | CCNB1, MAD2L1, CCNB2, BUB1, AURKA, CDC20 |
| hsa04110 | Cell cycle | 6 | 10.43 | 2.01E-06 | CCNB1, MAD2L1, CCNB2, BUB1, TTK, CDC20 | |
| hsa04115 | p53 signaling pathway | 4 | 6.95 | 2.36E-04 | CCNB1, CCNB2, RRM2, GTSE1 | |
| hsa04914 | Progesterone-mediated oocyte maturation pathway | 4 | 6.95 | 5.10E-04 | CCNB1, MAD2L1, CCNB2, BUB1 | |
| Module 2 | hsa04151 | PI3K-Akt signaling pathway | 4 | 25.82 | 0.00223 | VWF, TEK, ANGPT1, SPP1 |
| hsa04066 | HIF-1 signaling pathway | 3 | 19.37 | 0.00279 | EDN1, TEK, ANGPT1 | |
| Hubba Top 10 | hsa04512 | ECM-receptor interaction pathway | 4 | 40 | 3.80E-05 | VWF, CD36, ITGA8, COL1A1 |
| hsa04510 | Focal adhesion pathway | 4 | 40 | 4.95E-04 | VWF, CAV1, ITGA8, COL1A1 | |
| hsa04151 | PI3K-Akt signaling pathway | 3 | 30 | 0.03289 | VWF, ITGA8, COL1A1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes.
Expression and survival analysis of hub genes in NSCLC
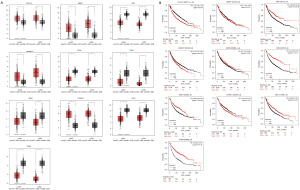
The GEPIA database was utilized to analyse the expression of the top 10 hub genes in NSCLC and adjacent lung tissues. COL1A1, MMP1, ADAM12 and CTHRC1 were significantly upregulated (P<0.05), whereas VWF, CD36, OGN, EDN1, CAV1 and ITGA8 were significantly downregulated (P<0.05) in NSCLC (Figure 4A). Survival curves were generated to assess the diagnostic efficiency of the 10 hub genes via Kaplan-Meier plotter. As shown in Figure 4B, two upregulated genes (COL1A1 and ADAM12) and five downregulated genes (VWF, OGN, EDN1, CAV1, and ITGA8) were significantly associated with survival prognosis in NSCLC (P<0.01).
Reanalysis of hub genes via KEGG enrichment analysis
KEGG enrichment analysis was reanalyzed to explore the possible pathways of the 7 hub genes via the DAVID database. Four genes (VWF, CAV1, ITGA8, COL1A1) were markedly enriched in the focal adhesion pathway (P=1.04E-04) and were upstream regulators of FAK (PTK2) (Table 5).
Table 5
| Pathway ID | Name | Count | % | P value | Genes |
|---|---|---|---|---|---|
| hsa04510 | Focal adhesion | 4 | 57.14 | 1.04E-04 | VWF, CAV1, ITGA8, COL1A1 |
| hsa04512 | ECM-receptor interaction | 3 | 42.86 | 9.33E-04 | VWF, ITGA8, COL1A1 |
| hsa04151 | PI3K-Akt signaling pathway | 3 | 42.86 | 0.01407 | VWF, ITGA8, COL1A1 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; NSCLC, non-small cell lung cancer.
Expression and survival analysis of PTK2 in NSCLC
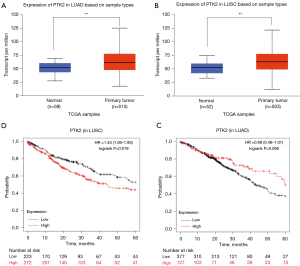
The expression of PTK2 between NSCLC and adjacent lung tissues was analysed using the UALCAN database. Compared to adjacent lung tissues, PTK2 was significantly overexpressed in both lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (P<0.01, Figure 5A,5B). PTK2 was associated with a poor prognosis in LUSC (HR =1.43, P<0.05, Figure 5C) but not in LUAD (HR =0.69, P>0.05, Figure 5D).
Discussion
NSCLC is the most common malignant cancer and has the fourth lowest survival rate of cancer types worldwide (19). Although numerous basic and clinical studies have been conducted to reveal the causes and underlying mechanisms of NSCLC formation and progression, the gene and mechanism of gene expression in NSCLC have not been systematically studied.
This study utilized bioinformatics methods to deeply analyse the four profile datasets downloaded from the GEO database. A total of 225 DEGs were obtained via the DAVID online database and classified into three groups (MF, BP and CC groups) by GO terms. The cell division GO term enriched both upregulated and downregulated DEGs, which indicates that the biological process of cell division may play vital roles in the development of NSCLC. For pathway analysis, upregulated DEGs were particularly enriched in oocyte meiosis, cell cycle and the p53 signaling pathway, while downregulated DEGs were enriched in malaria, vascular smooth muscle contraction and the PPAR signaling pathway (P<0.05). As a tumor inhibitor, p53 plays a pivotal role in cell biological functions, such as cell cycle progression, DNA damage response, apoptosis, senescence, and angiogenesis (20). Tac2-N (TC2N) acts as a novel oncogene by inhibiting apoptosis and promoting the proliferation of lung cancer cells by inhibiting the p53 signaling pathway (21).
On the basis of CytoHubba plugin of the Cytoscape software platform, the top 10 hub genes were identified, which were mainly involved in ECM-receptor interaction, focal adhesion and the PI3K-Akt signaling pathway. Four of the top 10 genes were enriched in the focal adhesion pathway and were upstream regulators of FAK (PTK2), including VWF, CAV1, ITGA8 and COL1A1. The integrin-activated adhesion kinase (FAK)-mediated signaling pathway is an important pathway in tumor invasion and metastasis (22). PTK2, as an adhesion protein kinase, plays an important role in the FAK-mediated signaling pathway, which includes the transduction of signals released from integrins and growth factor receptors (23-25). In the present study, PTK2 expression was significantly higher in NSCLC than in adjacent lung tissues. Moreover, PTK2 overexpression has been associated with poor survival in LUSC, while no correlation has been found in LUAD. As in our study, multiple studies demonstrated that PTK2 is overexpressed and/or activated in many tumor types, including NSCLC (26-30). It was previously reported that PTK2 overexpression evaluated by IHC has been correlated with worse overall survival (31,32). These above data illustrate that PTK2 may play an important role in tumour formation and progression. Therefore, small-molecule inhibitors targeting the PTK2 kinase domain have been undergoing preclinical and clinical investigation and induce cancer regression in solid cancers, including NSCLC (33-37). As a robust inhibition of PTK2, PF-562, 271 provides the potential to enhance cancer therapy with its novel target and dual antitumor and antiangiogenesis mechanisms of action and represents an unprecedented approach to NSCLC treatment through PTK2 inhibition (38).
Conclusions
In summary, 225 DEGs were identified in the current study. Among them, four hub genes were found, which were enriched in the focal adhesion pathway and were upstream regulators of PTK2. PTK2 may be a key oncogene leading to tumour formation and progression and associated with poor survival in NSCLC. However, clinical experiments are urgently needed to evaluate the molecular role of PTK2 in NSCLC. As a biomarker of NSCLC, the molecular mechanisms and clinical application of PTK2 require exploration in future studies.
Acknowledgments
We would like to thank PhD Lijia Xiao for his help in polishing this article.
Funding: This study was funded by Science and Technology Planning Project of Shenzhen (No. JCYJ20140415151845365) and Basic Medical and Health Research Project of Baoan District (No. 2020JD428).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1986/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1986/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-1986/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. Erratum in: CA Cancer J Clin 2011;61:134. [Crossref] [PubMed]
- Govindan R, Page N, Morgensztern D, et al. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol 2006;24:4539-44. [Crossref] [PubMed]
- Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: current status and perspectives. Crit Rev Oncol Hematol 2021;157:103194. [Crossref] [PubMed]
- Friedlaender A, Addeo A, Russo A, et al. Targeted therapies in early stage NSCLC: hype or hope? Int J Mol Sci 2020;21:6329. [Crossref] [PubMed]
- Vansteenkiste J, Crinò L, Dooms C, et al. 2nd ESMO consensus conference on lung cancer: early-stage non-small-cell lung cancer consensus on diagnosis, treatment and follow-up. Ann Oncol 2014;25:1462-74. [Crossref] [PubMed]
- Tsao AS, Scagliotti GV, Bunn PA Jr, et al. Scientific advances in lung cancer 2015. J Thorac Oncol 2016;11:613-38. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Fu Q, Yang F, Zhao J, et al. Bioinformatical identification of key pathways and genes in human hepatocellular carcinoma after CSN5 depletion. Cell Signal 2018;49:79-86. [Crossref] [PubMed]
- Gong L, Zhang D, Dong Y, et al. Integrated bioinformatics analysis for identificating the therapeutic targets of aspirin in small cell lung cancer. J Biomed Inform 2018;88:20-8. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Jiao X, Sherman BT. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics 2012;28:1805-6. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J 2021;19:4101-9. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- de Queiroz RM, Madan R, Chien J, et al. Changes in O-linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J Biol Chem 2016;291:18897-914. [Crossref] [PubMed]
- Hao XL, Han F, Zhang N, et al. TC2N, a novel oncogene, accelerates tumor progression by suppressing p53 signaling pathway in lung cancer. Cell Death Differ 2019;26:1235-50. [Crossref] [PubMed]
- Yan H, Guo M, Zou J, et al. Promotive effect of Talin-1 protein on gastric cancer progression through PTK2-PXN-VCL-E-Cadherin-CAPN2-MAPK1 signaling axis. J Clin Lab Anal 2020;34:e23555. [Crossref] [PubMed]
- Batista S, Maniati E, Reynolds LE, et al. Haematopoietic focal adhesion kinase deficiency alters haematopoietic homeostasis to drive tumour metastasis. Nat Commun 2014;5:5054. [Crossref] [PubMed]
- Sieg DJ, Hauck CR, Ilic D, et al. FAK integrates growth-factor and integrin signals to promote cell migration. Nat Cell Biol 2000;2:249-56. [Crossref] [PubMed]
- Zhao X, Guan JL. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv Drug Deliv Rev 2011;63:610-5. [Crossref] [PubMed]
- Gu HJ, Zhou B. Focal adhesion kinase promotes progression and predicts poor clinical outcomes in patients with osteosarcoma. Oncol Lett 2018;15:6225-32. [Crossref] [PubMed]
- Almstedt K, Sicking I, Battista MJ, et al. Prognostic significance of focal adhesion kinase in node-negative breast cancer. Breast Care (Basel) 2017;12:329-33. [Crossref] [PubMed]
- Omura G, Ando M, Saito Y, et al. Association of the upregulated expression of focal adhesion kinase with poor prognosis and tumor dissemination in hypopharyngeal cancer. Head Neck 2016;38:1164-9. [Crossref] [PubMed]
- Gómez Del Pulgar T, Cebrián A, Fernández-Aceñero MJ, et al. Focal adhesion kinase: predictor of tumour response and risk factor for recurrence after neoadjuvant chemoradiation in rectal cancer. J Cell Mol Med 2016;20:1729-36. [Crossref] [PubMed]
- Li M, Hong LI, Liao M, et al. Expression and clinical significance of focal adhesion kinase and adrenomedullin in epithelial ovarian cancer. Oncol Lett 2015;10:1003-7. [Crossref] [PubMed]
- Hsu NY, Chen CY, Hsu CP, et al. Prognostic significance of expression of nm23-H1 and focal adhesion kinase in non-small cell lung cancer. Oncol Rep 2007;18:81-5. [Crossref] [PubMed]
- Wang C, Yang R, Yue D, et al. Expression of FAK and PTEN in bronchioloalveolar carcinoma and lung adenocarcinoma. Lung 2009;187:104-9. [Crossref] [PubMed]
- Zhang H, Shao H, Golubovskaya VM, et al. Efficacy of focal adhesion kinase inhibition in non-small cell lung cancer with oncogenically activated MAPK pathways. Br J Cancer 2016;115:203-11. [Crossref] [PubMed]
- Infante JR, Camidge DR, Mileshkin LR, et al. Safety, pharmacokinetic, and pharmacodynamic phase I dose-escalation trial of PF-00562271, an inhibitor of focal adhesion kinase, in advanced solid tumors. J Clin Oncol 2012;30:1527-33. [Crossref] [PubMed]
- Jones SF, Siu LL, Bendell JC, et al. A phase I study of VS-6063, a second-generation focal adhesion kinase inhibitor, in patients with advanced solid tumors. Invest New Drugs 2015;33:1100-7. [Crossref] [PubMed]
- Shimizu T, Fukuoka K, Takeda M, et al. A first-in-Asian phase 1 study to evaluate safety, pharmacokinetics and clinical activity of VS-6063, a focal adhesion kinase (FAK) inhibitor in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol 2016;77:997-1003. [Crossref] [PubMed]
- Constanzo JD, Tang KJ, Rindhe S, et al. PIAS1-FAK interaction promotes the survival and progression of non-small cell lung cancer. Neoplasia 2016;18:282-93. Erratum in: Neoplasia 2016;18:457. [Crossref] [PubMed]
- Roberts WG, Ung E, Whalen P, et al. Antitumor activity and pharmacology of a selective focal adhesion kinase inhibitor, PF-562,271. Cancer Res 2008;68:1935-44. [Crossref] [PubMed]


