
Hypomethylation status of miR-657 promoter region as biomarker for diagnosis of hepatocellular carcinoma: a retrospective study
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common form of cancer and the third leading cause of cancer deaths worldwide (1). Although there are multiple therapeutic methods for HCC, including surgery, trans-hepatic arterial chemotherapy and embolization, targeted therapy, and immunotherapy, the overall survival rate remains poor for patients with intermediate and advanced stage disease, with a median survival of approximately 10 to 20 months (2). By contrast, for early-stage patients, the 5-year survival rate can be as high as 50–70% (3). The commonly used biomarker for clinical diagnosis of HCC is alpha-fetoprotein (AFP). But for early diagnosis, AFP shows low level of sensitivity (4). Therefore, it is necessary to find effective biomarkers for early diagnosis of HCC.
Epigenetic modification analysis has several advantages over somatic mutation analysis for cancer detection. First, epigenetic modifications have higher detection sensitivity, as there are always many methylated CpG sites within each targeted gene, and they can be detected in all genomic contexts, not only in coding regions (5). Second, epigenetic modifications can be detected in the early stages of cancer progression, whereas somatic mutations are always quantified at later stages (6). Third, epigenetic modifications are much more stable in both fluid and tissue specimens, which makes them more convenient and reliable for clinical use (7). The epigenetic modification of gene expression has an important role in HCC (8). As one of the most-studied epigenetic modification methods, DNA methylation of gene promoter regions occurs not only in HCC but also in premalignant conditions including chronic viral hepatitis B and cirrhosis of the liver (9). Thus, it could potentially be used for early diagnosis of HCC. Multiple studies have demonstrated the biomarker roles of gene promoter region methylation in HCC (10).
MicroRNAs are noncoding RNA molecules of approximately 23 nucleotides that play important parts in gene regulation in multiple cancers by binding to the messenger RNAs of protein-coding genes to direct their post-transcriptional repression (11). MiR-657 is a rarely reported microRNA that is associated with tumor promotion. MiR-657 was shown to inhibit apoptosis in hematological cancer cells of myeloid origin by targeting the endoplasmic reticulum stress signaling pathway (12). MiR-657 was also reported to play a critical part in lung tumor development by regulating transcriptional factors of Wilms tumor 1 (WT1) and ETS variant 1 (ETV1) (13). In HCC, miR-657 was found to be overexpressed in cancerous tissues compared with normal tissues of HCC patients. Overexpression of miR-657 could enhance the proliferation and colony formation ability of HCC cells in vitro and induce tumor development in immunodeficient mice by directly targeting the 3’ untranslated region (UTR) of transducin-like enhancer protein 1 (TLE1) and activating nuclear factor kappa B (NF-κB) pathways (14). However, the methylation status of the miR-657 promoter region in cancers, especially in HCC, has remained unclear.
Here, we report for the first time that the methylation level of the miR-657 promoter region is decreased in cancerous tissues compared with normal tissues in HCC. Receiver operating characteristic (ROC) curve analysis revealed that the methylation level of the miR-657 promoter region could be used to effectively distinguish cancerous tissues from paired normal tissues in both AFP-positive and AFP-negative groups, indicating its potential as a supplementary biomarker for diagnosis of HCC.
We present the following article in accordance with the STARD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2621/rc).
Methods
Patient samples and clinical data collection
From July 2008 to February 2014, we collected human primary cancerous tissues and paired normal tissues from 160 HCC patients who were diagnosed and treated at the Zhejiang Cancer Hospital and Sir Run Run Shaw Hospital. Our study was conducted in strict compliance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committees of Zhejiang Cancer Hospital and Sir Run Run Shaw Hospital. The inclusion criteria were patients who diagnosed with HCC by histopathology and received curative surgery. The exclusion criteria were patients who had history of other cancers. Clinical data were obtained after each patient provided written informed consent for treatment.
Gene bioinformatics and primer design
Sequence information for the miR-657 gene was obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene). The Database of CpG Islands (http://dbcat.cgm.ntu.edu.tw/) was used to scan the sequence from 5,000 base pairs (bp) upstream to 5,000 bp downstream of the transcription start site (TSS) of miR-657. A candidate CpG island near the TSS was identified. The amplicon studied in our research was located on chr17:81124276-81127373. Fourteen CpG sites were found in our selected amplicon.
DNA extraction and bisulfite conversion
A QIAamp DNAMini Kit (QIAGEN, Hilden, Germany) was used to extract genomic DNA from ≥25 mg primary cancerous tissues and paired normal tissues. The concentrations and quality of all DNA samples were detected using a Thermo NanoDrop 2000 spectrophotometer. Then, 400–500 ng of DNA was taken for bisulfite treatment using an EpiTect Fast DNA Bisulfite Kit (QIAGEN, Hilden, Germany).
Methylation-specific PCR
The prepared DNA samples were amplified by PCR following SAP cleanup, T cleavage, and Clean Resin steps. Then, a MassARRAY Analyzer 4 (Sequenom, USA) was used for methylation analysis of each CpG site. The primers for methylation-specific PCR were as follows: forward, aggaagagagGTTAGGATGTTTAGTTTTTGGGGAT; reverse, cagtaatacgactcactatagggagaaggctCTCCAACCCAACCTACCTTAACTAC.
Statistical analysis
Sample size was calculated by PASS 15. The procedure of Confidence Intervals for One Proportion in PASS 15 software was used. Confidence level was 0.95. Confidence interval width was 0.1. Proportion was 0.9. The result showed that reasonable sample size was 158. So, we chose 160 as our sample size. Paired two-tailed t-tests were used to compare the differences in methylation of the miR-657 promoter region between cancerous tissues and paired normal tissues. Independent-samples t-test was used to compare the differences in methylation of the miR-657 promoter region between early and late stage HCC patients. Independent sample t-tests were used to investigate the relationships between methylation levels of the miR-657 promoter region and clinicopathological features of HCC patients. The correlations between methylation levels at each CpG site were assessed by Pearson correlation analysis. ROC curve analysis was used to evaluate the effectiveness of methylation levels as predictive biomarkers for diagnosis of HCC. Area under the curve (AUC) of greater than 0.8 was considered predictive. The diagnostic sensitivity, specificity and Youden’s index [sensitivity (%)+specificity (%) – 100] were determined from optimal AUC of methylation levels. A two-tailed P value less than 0.05 was considered significant. All data were analyzed using SPSS version 21.0.
Results
Patient features
The clinical features of the 160 HCC patients are listed in Table 1. There were 131 males and 29 females. The median age of all patients at diagnosis was 55 years. A total of 136 patients were infected with hepatitis B virus, of which only 19 received antiviral treatment. Twenty patients had a family history of HCC, and 105 patients had an alcohol habit (≥50 mg/day). AFP levels were positive in 53 patients, of which 52 had an AFP level higher than 400 ng/mL. One-hundred patients had liver cirrhosis and 47 patients were classified as BCLC B–D stage. Regarding pathological characteristics, 121 patients were classified as stage I–II according to the International Cancer Control TNM staging system.
Table 1
| Parameters | Categories | n |
|---|---|---|
| Gender | Female | 29 |
| Male | 131 | |
| Age | <55 | 81 |
| ≥55 | 79 | |
| Age (mean ± SD) | 54.0±12.1 | |
| Hypertension | No | 123 |
| Yes | 37 | |
| Type 2 diabetes | No | 140 |
| Yes | 20 | |
| Family history | No | 140 |
| YES | 20 | |
| Alcohol habit (≥50 mg/day) | No | 55 |
| Yes | 105 | |
| HBV infection | No | 22 |
| Yes | 136 | |
| Preoperative anti-HBV treatment | No | 141 |
| Yes | 19 | |
| Liver cirrhosis stage | 0 | 13 |
| 1 | 38 | |
| 2 | 51 | |
| 3 | 11 | |
| AFP | Negative | 104 |
| Positive | 53 | |
| AFP level | AFP ≤400 ng/mL | 96 |
| AFP >400 ng/mL | 52 | |
| AFP (mean ± SE) | 2,711.14±578.14 ng/mL | |
| ALT (mean ± SE) | 45.08±2.54 U/L | |
| AST (mean ± SE) | 44.93±2.50 U/L | |
| ALP (mean ± SE) | 95.84±3.92 U/L | |
| γ-GT (mean ± SE) | 91.05±9.52 U/L | |
| TBIL (mean ± SE) | 16.86±1.00 μmol/L | |
| DBIL (mean ± SE) | 5.68±0.23 μmol/L | |
| IBIL (mean ± SE) | 10.38±0.41 μmol/L | |
| BCLC stage | A | 112 |
| B-D | 47 | |
| TMN stage | I-II | 121 |
| III-IV | 38 | |
HBV, Hepatitis B Virus; AFP, alpha-fetoprotein; ALT, alanine transaminase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; γ-GT, gamma-glutamyltransferase; TBIL, total Bilirubin; DBIL, direct bilirubin; IBIL, indirect bilirubin; BCLC, Barcelona clinic liver cancer; TMN, tumor node metastasis.
Methylation levels of miR-657 promoter region in cancerous tissues and paired normal tissues
Fourteen CpG sites were identified in our selected amplicon; 12 of these (CpG1, CpG2, CpG3.4, CpG5.6, CpG7, CpG8, CpG9, CpG10, CpG13, CpG14) were genotyped successfully (Figure 1). CpG3 and CpG4 were genotyped as CpG3.4 because they were very close to each other. For the same reason, CpG5 and CpG6 were also genotyped as CpG5.6.
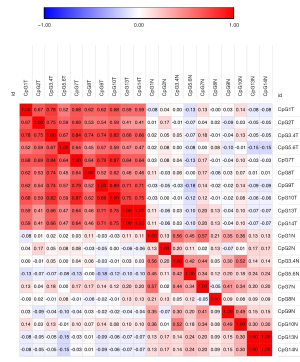
Hypomethylation of each CpG site in the miR-657 promoter region was detected in cancerous tissues compared with paired normal tissues (Table 2, Figure 2). The mean methylation level of the miR-657 promoter region was also significantly lower in cancerous tissues than in paired normal tissues (48.91%:67.04%, P<0.0001, Table 2). Correlation analysis revealed that the methylation levels at different CpG sites were significantly associated with each other (Figure 3).
Table 2
| CpGs | Group | Mean% | ⊿Mean% | P value |
|---|---|---|---|---|
| CpG1 | Tumor | 52.40 | 27.56 | <0.0001 |
| Normal | 79.96 | |||
| CpG2 | Tumor | 34.12 | 5.37 | <0.0001 |
| Normal | 39.49 | |||
| CpG3.4 | Tumor | 64.68 | 23.28 | <0.0001 |
| Normal | 87.96 | |||
| CpG5.6 | Tumor | 67.92 | 14.71 | <0.0001 |
| Normal | 82.63 | |||
| CpG7 | Tumor | 47.15 | 15.61 | <0.0001 |
| Normal | 62.76 | |||
| CpG8 | Tumor | 59.27 | 18.48 | <0.0001 |
| Normal | 77.75 | |||
| CpG9 | Tumor | 35.36 | 22.38 | <0.0001 |
| Normal | 57.74 | |||
| CpG10 | Tumor | 47.63 | 26.26 | <0.0001 |
| Normal | 73.89 | |||
| CpG13 | Tumor | 41.64 | 13.14 | <0.0001 |
| Normal | 54.78 | |||
| CpG14 | Tumor | 41.64 | 13.14 | <0.0001 |
| Normal | 54.87 | |||
| Mean | Tumor | 48.91 | 18.13 | <0.0001 |
| Normal | 67.04 |
We also calculated the methylation level of each CpG site in miR-657 promoter region for different stage of HCC patients. The result showed that there was no difference for methylation level of miR-657 promoter region in early and late stage of HCC patients (0.4897:0.4850, P=0.071, Table 3).
Table 3
| CpGs | HCC stage | Mean% | P value |
|---|---|---|---|
| CpG1 tumor | Early HCC | 0.5139 | 0.548 |
| Late HCC | 0.5524 | ||
| CpG2 tumor | Early HCC | 0.3333 | 0.734 |
| Late HCC | 0.3673 | ||
| CpG3.4 tumor | Early HCC | 0.6430 | 0.168 |
| Late HCC | 0.6615 | ||
| CpG5.6 tumor | Early HCC | 0.6823 | 0.853 |
| Late HCC | 0.6595 | ||
| CpG7 tumor | Early HCC | 0.4665 | 0.712 |
| Late HCC | 0.4817 | ||
| CpG8 tumor | Early HCC | 0.5726 | 0.542 |
| Late HCC | 0.6190 | ||
| CpG9 tumor | Early HCC | 0.3634 | 0.094 |
| Late HCC | 0.3100 | ||
| CpG10 tumor | Early HCC | 0.4838 | 0.286 |
| Late HCC | 0.4307 | ||
| CpG13 tumor | Early HCC | 0.4188 | 0.129 |
| Late HCC | 0.3841 | ||
| CpG14 tumor | Early HCC | 0.4188 | 0.129 |
| Late HCC | 0.3841 | ||
| Mean tumor | Early HCC | 0.4897 | 0.071 |
| Late HCC | 0.4850 |
HCC, hepatocellular carcinoma.
Correlation between methylation levels of miR-657 promoter region and AFP levels in HCC patients
To explore whether the hypomethylation of miR-657 promotor region had a role in HCC, we evaluated the association between methylation levels of the miR-657 promotor region and clinical features of HCC patients. The results showed that methylation levels of the miR-657 promotor region were associated with AFP levels in HCC patients. The methylation levels of two CpG sites (CpG2 tumor and CpG8 tumor) and the mean methylation level of the miR-657 promoter region in cancerous tissues were significantly associated with AFP levels in HCC patients (Table 4). The mean methylation level of the miR-657 promoter region in AFP-positive HCC patients was 46.76%, which was significantly lower than that in AFP-negative HCC patients (53.02%, P=0.043, Table 4). The differences in methylation levels of four CpG sites between tumor tissues and paired normal tissues (CpG7, CpG9, CpG13, and CpG14) and the differences in mean methylation levels of the miR-657 promoter region were also significantly associated with AFP levels of HCC patients (Table 4).
Table 4
| CpG sites | AFP | Mean methylation level (%) | P value |
|---|---|---|---|
| CpG2 tumor | Yes | 32.99 | 0.048 |
| No | 37.07 | ||
| CpG8 tumor | Yes | 54.53 | 0.014 |
| No | 67.27 | ||
| Mean tumor | Yes | 46.76 | 0.043 |
| No | 53.02 | ||
| CpG7 difference | Yes | 18.32 | 0.018 |
| No | 10.26 | ||
| CpG9 difference | Yes | 26.21 | 0.008 |
| No | 14.82 | ||
| CpG13 difference | Yes | 16.45 | 0.025 |
| No | 6.61 | ||
| CpG14 difference | Yes | 16.45 | 0.025 |
| No | 6.61 | ||
| Mean difference | Yes | 20.99 | 0.012 |
| No | 12.11 |
Difference represents the difference of methylation level between tumor tissue and paired normal tissue. AFP, alpha-fetoprotein.
ROC curve analysis of miR-657 promoter region methylation levels in cancerous tissues and paired normal tissues of HCC patients
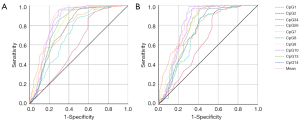
As the promoter region of miR-657 was significantly hypomethylated in cancerous tissues compared with paired normal tissues, and its methylation levels were associated with AFP levels in HCC patients, we further calculated the AUC of the ROC curves to determine whether the methylation status of the miR-657 promoter region could be used as a biomarker for diagnosis of HCC. The results showed that the methylation levels of 5 CpG sites (CpG1, CpG34, CpG7, CpG9, CpG10) and the mean methylation level of the miR-657 promoter region could effectively distinguish cancerous tissues from paired normal tissues, with the mean methylation level being the most powerful indicator (AUC =0.847, P<0.001; Table 5, Figure 4A). Therefore, we chose the mean methylation level for further analysis. ROC analysis indicated that the optimal cut-off value for the mean methylation level of miR-657 was 59.50%, with a sensitivity of 95.50% and specificity of 70.01%. We further checked the effectiveness of the mean methylation level for diagnosis of HCC in early-stage HCC patients (BCLC A). The results showed that the methylation levels of 3 CpG sites (CpG1, CpG 7, CpG 10) and the mean methylation level of the miR-657 promoter region could effectively distinguish cancerous tissues from paired normal tissues (Table 5, Figure 4B). With a cut-off value of 60.20%, the mean methylation level could distinguish cancerous tissues from paired normal tissues with a sensitivity of 93.80% and specificity of 69.96%.
Table 5
| CpG sites | All patients | Early HCC | AFP (−) | AFP (+) | AFP ≤400 ng/mL | AFP >400 ng/mL | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUC | P | AUC | P | AUC | P | AUC | P | AUC | P | AUC | P | ||||||
| CpG1 | 0.815 | <0.001 | 0.825 | <0.001 | 0.771 | <0.001 | 0.831 | <0.001 | 0.783 | <0.001 | 0.862 | <0.001 | |||||
| CpG2 | 0.601 | 0.004 | 0.643 | 0.001 | 0.526 | 0.674 | 0.638 | 0.001 | 0.572 | 0.114 | 0.645 | 0.020 | |||||
| CpG3.4 | 0.801 | <0.001 | 0.773 | <0.001 | 0.720 | <0.001 | 0.837 | <0.001 | 0.758 | <0.001 | 0.860 | <0.001 | |||||
| CpG5.6 | 0.785 | <0.001 | 0.786 | <0.001 | 0.715 | 0.001 | 0.817 | <0.001 | 0.772 | <0.001 | 0.785 | <0.001 | |||||
| CpG7 | 0.801 | <0.001 | 0.802 | <0.001 | 0.716 | 0.001 | 0.843 | <0.001 | 0.781 | <0.001 | 0.856 | <0.001 | |||||
| CpG8 | 0.706 | <0.001 | 0.724 | <0.001 | 0.644 | 0.021 | 0.739 | <0.001 | 0.679 | <0.001 | 0.718 | <0.001 | |||||
| CpG9 | 0.838 | <0.001 | 0.798 | <0.001 | 0.751 | <0.001 | 0.874 | <0.001 | 0.820 | <0.001 | 0.845 | <0.001 | |||||
| CpG10 | 0.834 | <0.001 | 0.803 | <0.001 | 0.755 | <0.001 | 0.865 | <0.001 | 0.801 | <0.001 | 0.872 | <0.001 | |||||
| CpG13 | 0.763 | <0.001 | 0.741 | <0.001 | 0.671 | 0.006 | 0.806 | <0.001 | 0.747 | <0.001 | 0.784 | <0.001 | |||||
| CpG14 | 0.763 | <0.001 | 0.741 | <0.001 | 0.671 | 0.006 | 0.806 | <0.001 | 0.747 | <0.001 | 0.784 | <0.001 | |||||
| Mean | 0.847 | <0.001 | 0.822 | <0.001 | 0.712 | 0.001 | 0.898 | <0.001 | 0.794 | <0.001 | 0.909 | <0.001 | |||||
HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; AUC, area under the curve.
Discussion
In this study, we first reported that the methylation level of the miR-657 promoter region was significantly lower in cancerous tissues than in paired normal tissues of HCC patients. We calculated the AUC of ROC curves for all 160 patients to determine whether the methylation level of miR-657 could be used as a biomarker for diagnosis of HCC. The results showed that the methylation level of the miR-657 promoter region could effectively distinguish cancerous tissues from paired normal tissues of HCC patients. AFP is the most widely used biomarker for diagnosis of HCC, with a sensitivity of 61% and specificity of 86% when using a cut-off value of 20–100 ng/mL (15). When the cut-off value was increased to 400 ng/mL, the specificity increased to 99%, but the sensitivity fell to 32% (16). In our study, when using a cut-off value of 59.50%, the mean methylation level of miR-657 could distinguish cancerous tissues from paired normal tissues with a sensitivity of 95.50% and specificity of 70.01%. We also checked the effectiveness of the mean methylation level for diagnosis of early HCC. It was previously reported that an AFP value of 20 ng/mL could be used for diagnosis of early-stage HCC (stage BCLC A) with a sensitivity of 53% and specificity of 90% (4). In our study, when using a cut-off value of 60.2%, the mean methylation level of miR-657 could distinguish cancerous tissues from paired normal tissues with a sensitivity of 93.80% and specificity of 69.96% in the early HCC (stage BCLC A) subgroup. Therefore, we concluded that the methylation level of the miR-657 promoter region could be used as an alternative and supplementary biomarker for diagnosis of HCC, with much higher sensitivity and comparable specificity compared to serum AFP levels.
Our study revealed that methylation levels of the miR-657 promoter region were associated with serum AFP levels in HCC patients. The methylation levels of the miR-657 promoter region were significantly lower in the AFP-positive group than in the AFP-negative group. AFP is the most widely used biomarker for diagnosis of HCC and has been reported to be correlated with histologic differentiation of HCC, with higher AFP levels in poorly differentiated HCC than in moderately differentiated HCC (17). miR-657 has been shown to promote hepatocarcinogenesis by directly targeting the TLE1 3’ UTR and activating NF-κB pathways, and overexpression of miR-657 significantly promoted the spheroid formation of hepatoma cancer cells in vitro (14). Thus, we propose that miR-657 may promote hepatocarcinogenesis by regulating the differentiation of hepatoma cancer cells.
Our study had some limitations. First, we only measured the methylation levels of the miR-657 promoter region in HCC patients; the expression of miR-657 was not measured. Thus, whether overexpression of miR-657 was caused by the demethylation of miR-657 promoter region remains unclear. Further studies are needed to explore the effects of methylation on expression of miR-657. Second, the methylation levels of miR-657 promoter region in serum of HCC patients were not measured. If alterations in methylation levels of miR-657 collected from serum could effectively distinguish HCC patients from a normal population, they could be more widely used in clinical settings as biomarkers of HCC.
Conclusions
Our study revealed that methylation levels of miR-657 were decreased in HCC patients and could be used as an alternative and supplementary biomarker for diagnosis of HCC.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2621/rc
Data Sharing Statement: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2621/dss
Peer Review File: Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2621/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2621/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet 2018;391:1301-14. [Crossref] [PubMed]
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol 2017;34:153-9. [Crossref] [PubMed]
- Duvoux C, Roudot-Thoraval F, Decaens T, et al. Liver transplantation for hepatocellular carcinoma: a model including α-fetoprotein improves the performance of Milan criteria. Gastroenterology 2012;143:986-94.e3; quiz e14-5. [Crossref] [PubMed]
- Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet 2019;20:109-27. [Crossref] [PubMed]
- Lorincz AT. The Promise and the Problems of Epigenetics Biomarkers in Cancer. Expert Opin Med Diagn 2011;5:375-9. [Crossref] [PubMed]
- García-Giménez JL, Mena-Mollá S, Beltrán-García J, et al. Challenges in the analysis of epigenetic biomarkers in clinical samples. Clin Chem Lab Med 2017;55:1474-7. [Crossref] [PubMed]
- Nakamura M, Chiba T, Kanayama K, et al. Epigenetic dysregulation in hepatocellular carcinoma: an up-to-date review. Hepatol Res 2019;49:3-13. [Crossref] [PubMed]
- Tischoff I, Tannapfe A. DNA methylation in hepatocellular carcinoma. World J Gastroenterol 2008;14:1741-8. [Crossref] [PubMed]
- Zhang C, Li J, Huang T, et al. Meta-analysis of DNA methylation biomarkers in hepatocellular carcinoma. Oncotarget 2016;7:81255-67. [Crossref] [PubMed]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215-33. [Crossref] [PubMed]
- Lim HJ, Park MN, Kim C, et al. MiR-657/ATF2 Signaling Pathway Has a Critical Role in Spatholobus suberectus Dunn Extract-Induced Apoptosis in U266 and U937 Cells. Cancers (Basel) 2019;11. [Crossref] [PubMed]
- Jin X, Guan Y, Sheng H, et al. Crosstalk in competing endogenous RNA network reveals the complex molecular mechanism underlying lung cancer. Oncotarget 2017;8:91270-80. [Crossref] [PubMed]
- Zhang L, Yang L, Liu X, et al. MicroRNA-657 promotes tumorigenesis in hepatocellular carcinoma by targeting transducin-like enhancer protein 1 through nuclear factor kappa B pathways. Hepatology 2013;57:1919-30. [Crossref] [PubMed]
- Zhang J, Chen G, Zhang P, et al. The threshold of alpha-fetoprotein (AFP) for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. PLoS One 2020;15:e0228857. [Crossref] [PubMed]
- Marrero JA, Feng Z, Wang Y, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology 2009;137:110-8. [Crossref] [PubMed]
- Shan Q, Chen J, Zhang T, et al. Evaluating histologic differentiation of hepatitis B virus-related hepatocellular carcinoma using intravoxel incoherent motion and AFP levels alone and in combination. Abdom Radiol (NY) 2017;42:2079-88. [Crossref] [PubMed]




