
High expression of TTC21A predict poor prognosis of colorectal cancer and influence the immune infiltrating level
Introduction
Colorectal cancer (CRC) is one of the most common malignancies worldwide, and it’s also the second most common cause of cancer deaths. Meanwhile, it accounts for 8–9% of cancer-related mortality, with poor 5-year survival for stage IV disease (1-4). CRC ranked the third highest in the incidence rate in China, while the mortality rate ranked fifth. Approximately 20% of patients with CRC have metastatic disease at diagnosis. Despite improvements in systemic treatments for patients with metastatic disease, the 5-year survival rate is only 12–14% in patients with metastatic CRC (2,5,6).
Current management strategies for CRC treatment mainly include surgery, chemotherapy, radiotherapy, and immunotherapy (7,8). Although immunotherapy improves the prognosis of CRC, there are still a series of problems (9), such as individual differences, conspicuously reducing patients’ quality of life, and adverse immune reactions (10,11). The mechanism underlying CRC non-response to ICI immunotherapy is currently highly debated (8). At present, staging is a reference standard for the prognosis of CRC (12), but methods to improve the predictive value of survival in CRC patients are needed. Thus, further research is necessary for CRC treatment, such as studying the immunophenotypes of tumor-immune communications, identifying intelligent biomarkers, and exclusive immune-related therapeutic targets.
The predictive and prognostic significance of tumor-infiltrating lymphocytes (TILs) has been highlighted in various solid cancers such as melanoma, lung cancer, and CRC (13-15). Galon and colleagues first demonstrated the positive prognostic impact of TILs in CRC (16). CD8+ TILs are good prognostic factors for CRC. Th1 cells have an essential role in initiating and maintaining an effective CD8+ cytotoxic T cell response, recruitment of CD8+ cells to the tumor bed, and indirectly mediating immunological tumor cell death (16-18). Thus, TILs reflect the host antitumor immune response, and their density and localization were prognostic in patients with CRC (18,19). Nonetheless, the traditional prognostic association of immune infiltration in CRC remains unsure.
TTC21A (tetratricopeptide repeat domain 21A), encodes proteins in the tetratricopeptide repeat (TPR) family and is expressed in various tissues and organs (20,21). TPRs are a widely studied structural motif that mediates intermolecular protein interactions and facilitates interactions with a variety of ligands or substrates (22). Interestingly, although the molecular function of TTC21A is unknown, data from the functional protein association networks (STRING) indicate that the vast majority of the predicted functional partners of human TTC21A and mouse TTC21A are IFT components. Research by Liu and his colleagues also confirmed that TTC21A is likely an IFT component interacting with IFT proteins (23). Furthermore, there are an increasing number of reports linking IFT protein dysfunction to different types of malignancies (24-27). However, there square measure few reports on the involvement of TTC21A in tumorigenesis and progression. Only one report by Wang et al. confirmed the association between TTC21A and lung adenocarcinoma (28). This study downloaded somatic mutations and RNA-SEQ data from CRC patients from The Cancer Genome Atlas (TCGA) database. Then, the correlation between TTC21A and CRC prognosis was analyzed using R statistical software 3.6.3 with the “ggplot2 (3.3.3)”, “survival (3.2-10)”, “rms (6.2-0)” packages.
Moreover, the relative proportion of different types of tumor-infiltrating immune cells (TIICs) in different tumor microenvironments was detected by the “GSVA” package, and the relationship between TTC21A and TIICs was studied (1). The findings of this study deepen our understanding of the possible role of TTC21A in CRC and shed light on the potential relevance and possible mechanisms of TTC21A’s interaction with tumor immunity. A flow chart summarizing our work is shown in Figure 1.
Therefore, TTC21A may be a new predictor of prognosis and immune infiltration in CRC patients. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2674/rc).
Methods
Data acquisition
A dataset of patients with CRC containing gene expression profiles and paired clinical information was downloaded from the publicly available TCGA database (https://portal.gdc.cancer.gov), including 480 tumor tissues, excluding cases with insufficient or missing data on local invasion, lymph node metastasis, distant metastasis, age, overall survival, and TNM staging. Finally, Cox regression analysis was performed on 477 eligible clinical data. Furthermore, to investigate the effect of TTC21A expression on the immune microenvironment, 454 tumor tissues were collected for ssGSEA analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Survival and expression analysis
The “ggplot2 (3.3.3)”, “survival (3.2-10)” and “rms (6.2-0)” software packages of R statistical software 3.6.3 were used to evaluate the correlation between the expression of TTC21A and the clinicopathological information of CRC. At the same time, a boxplot with tumor or normal as a variable was drawn to visually display the differential TTC21A and TTC21B expression in tumor and normal tissue. The pathological stage was used as the variable to draw the clinical-stage boxplot and compare the expression of TTC21A in various pathological stages.
TTC21A expression in tumor tissues and cells
The Human Protein Atlas (HPA) (https://www.proteinatlas.org) was applied to show the TTC21A expression in protein level. In addition, the Human Colon Cancer Atlas (https://singlecell.broadinstitute.org/single_cell) and Deeply Integrated Human Single-Cell Omics (DISCO) data (https://www.immunesinglecell.org/genepage/TTC21A) were further used to analysis the gene expression in CRC cells.
Evaluation of TIICs
We used 521 CRC samples from TCGA containing all genes to assess the connection between TTC21A expression and TIICs. Lymphocytes that will be affected by TTC21A expression were designated with a P value <0.05 as the criterion.
Single-sample GSEA, a modification of standard GSEA, was performed on RNA measurements for each sample using the “GSVA” package in R version 3.6.3. We plotted the correlated lollipops to detect the correlation between TTC21A and 24 types of immune cells.
In addition, we used ssGSEA to plot the correlation expression scatter plot to confirm the relationship between TTC21A expression and TIICs, plotted together with Spearman’s R and predicted statistical significance. P value <0.05.
The immune cells included: activated dendritic cells (aDC), B cells, CD8 T cells, Cytotoxic cells, dendritic cells (DC), eosinophils, immature DC (iDC), macrophages, mast cells, neutrophils, NK CD56 bright cells, NK CD56 dim cells, NK cells, plasmacytoid DC (pDC), T cells, T helper cells, T central memory (Tcm), T effector memory (Tem), T follicular helper (Tfh), T gamma delta (Tgd), Th1 cells, Th17 cells, Th2 cells, and Treg (1,29). SsGSEA helped map immune cell box plots with different TTC21A expressions and Spearman’s R and predicted statistical significance. P value <0.05 indicates the threshold.
Statistical analysis
R-3.6.3 processed all the statistical data obtained by TCGA. Logistic regression was used to analyze the correlation between TTC21A expression and clinical features. At the same time, Cox regression analysis was used to determine PATIENTS’ OS-related clinical characteristics with TCGA. Spearman’s R method and statistical significance were used to analyze the correlation of gene expression. The absolute value of R is greater than 0.1, and the value of P<0.05 is considered statistically significant.
Results
Gene expression of TTC21A and TTC21B
Analysis of the TCGA cohort revealed that gene expression of TTC21A and TTC21B differ from normal tissue in many tumor tissues (Figure 2A,2B), both of the TTC21A and TTC21B expression were found to be up-regulated in CRC tissues compared with the normal tissues (Figure 2C; Log2FC <2, P<0.05).
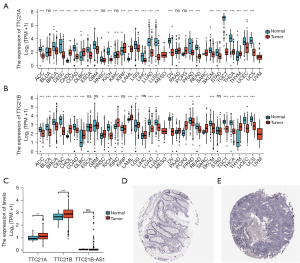
We further applied the HPA database to explore the protein expression of TTC21A in CRC and found that the staining was negative in tumor tissues, and there was staining protein expression in interstitial tumor tissues. These findings propose that the effect of TTC21A on CRC may be influenced by matrix components (Figure 2D,2E).
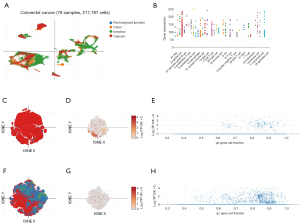
We also investigated the distribution of TTC21A in CRC cells in detail. Using DISCO data (https://www.immunesinglecell.org), we found that TTC21A was predominantly expressed in germinal center B cell, Treg, stromal cell, smooth muscle cell, mast cell, CD8 T cell, and NK T cell (Figure 3A,3B). We applied the Single Cell Portal (https://singlecell.broadinstitute.org/single_cell) and found TTC21A was mainly expressed in immune cells and T/NK/ILC cells (Figure 3C-3H). These results indicate that TTC21A impacts CRC maybe by modifying immune cell infiltration.
Survival outcomes and multivariate analysis
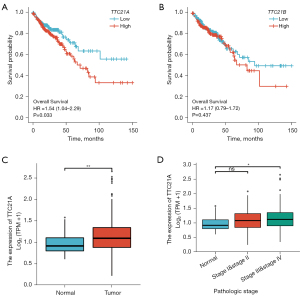
Survival analyses for TCGA CRC patients demonstrated that tumor expression of TTC21A was associated with shorter overall survival (Figure 4A, P=0.033) while TTC21B was not correlated with the prognosis (Figure 4B). Additional analyses showed that TTC21A expression was higher in CRC samples than in normal samples (Figure 4C, Log2FC <2, P value <0.05), whereas increased TTC21A expression was significantly associated with an advanced pathological stage (Figure 4D, P<0.05).
And then, as shown in Figure 5A and Table 1, univariate analysis using Cox regression showed that several factors, including age (HR =0.621; P=0.028), tumor status (HR =0.325, P=0.004), distant metastasis (HR =0.238, P<0.001), lymph node status (HR =0.386, P<0.001), and TTC21A expression (HR =0.649, P=0.033) were significantly associated with overall survival.
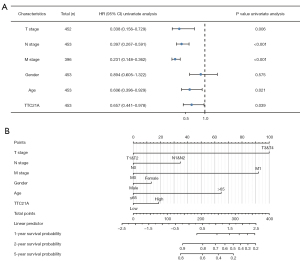
Table 1
| Characteristics | Total (n) | Univariate analysis | |
|---|---|---|---|
| HR (95% CI) | P value | ||
| T stage (tumor status) | 476 | 0.325 (0.151–0.703) | 0.004 |
| N stage (lymph node) | 477 | 0.386 (0.259–0.574) | <0.001 |
| M stage (distant metastasis) | 414 | 0.238 (0.153–0.373) | <0.001 |
| Age (≤65 vs. >65) | 477 | 0.621 (0.406–0.951) | 0.028 |
| Gender (female vs. male) | 477 | 0.908 (0.615–1.340) | 0.627 |
| TTC21A (low vs. high) | 477 | 0.649 (0.437–0.966) | 0.033 |
In the multivariate analysis of the “rms” package analysis (30), independent prognostic factors for CRC included upregulation of TTC21A expression, high tumor stage, and distant metastasis (Figure 5B).
TTC21A expression and clinicopathologic changes
The expression mechanism of TTC21A in tumors must be more studied. Therefore, we tend to analyze the link between TTC21A and some clinical aspects of CRC samples. CRC cases with qualified clinical data were investigated using R-3.6.3 software.
As shown in Table 2, the logistic multivariate analysis with TTC21A expression because the classification variable (median expression value is 2.5) showed that inflated TTC21A expression was related to the pathological stage (I&II&III vs. IV, P=0.557), tumor status (T3&T4 vs. T1&T2, P=0.662), and lymph nodes (N1&N2 vs. N0, P=0.193).
Table 2
| Characteristics | Total (n) | OR (95% CI) | P value |
|---|---|---|---|
| T stage (T3&T4 vs. T1&T2) | 477 | 1.106 (0.704–1.741) | 0.662 |
| N stage (N1&N2 vs. N0) | 478 | 1.275 (0.885–1.841) | 0.193 |
| M stage (M1 vs. M0) | 415 | 1.203 (0.710–2.045) | 0.493 |
| Age (>65 vs. ≥65) | 478 | 1.035 (0.718–1.492) | 0.852 |
| Gender (male vs. female) | 478 | 1.000 (0.698–1.432) | 1.000 |
| Pathologic stage (stage I&stage II&stage III vs. stage IV) | 467 | 0.855 (0.506–1.441) | 0.557 |
TTC21A expression and TIICs
Preceding studies have shown that TILs are independent prognostic factors for sentinel lymph node status and survival in patients with malignant tumors (31). Thus, we attempted to find a relationship between TTC21A expression and immune infiltration in CRC. We analyzed the RNA-SEQ data of 454 TCGA samples and included the expression of the first half and the last half of TTC21A into the high and low expression groups, respectively. Then, infiltration levels of each of the 24 immune cell types were measured using ssGSEA. Finally, 227 patients in the high expression group and 227 in the low expression group met the screening criteria. The results were shown in Figure 6A-6C. The proportions of the 24 subgroups of immune cells were clearly shown above.
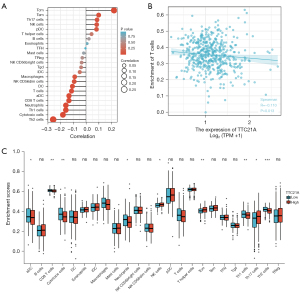
Figure 6A,6C show that the primary immune cells affected by TTC21A expression were CD8 T cells, cytotoxic cells, neutrophils, Th1 cells, Th2 cells, NK cells, T central memory cells, and Th17 cells. Among them, the proportion of CD8 T cells (P=0.004), cytotoxic cells (P=0.001), neutrophils (P=0.009), Th1 cells (P=0.009) and Th2 cells (P<0.001) in the high expression group was higher than that in the low expression group.
Moreover, the proportions of NK cells (P=0.042), T central memory (P=0.002), and Th17 cells (P=0.015) were significantly decreased. In addition, a scatter plot (Figure 6B) showed a negative correlation between CD8+ T cell infiltration and TTC21A expression in this cohort of CRC patients. Spearman correlation coefficient was used to evaluate the correlation. TTC21A was associated with similar results for neutrophils and NK cell markers (Table 3). Therefore, these findings suggest that TTC21A may play an essential role in regulating the abundance of T cells, neutrophils, and NK cells.
Table 3
| Gene | Cells | R (Pearson) | P (Pearson) | R (Spearman) | P (Spearman) |
|---|---|---|---|---|---|
| TTC21A | aDC | −0.158 | <0.001 | −0.133 | 0.003 |
| TTC21A | B cells | 0.036 | 0.429 | 0.023 | 0.613 |
| TTC21A | CD8 T cells | −0.194 | <0.001 | −0.134 | 0.003 |
| TTC21A | Cytotoxic cells | −0.181 | <0.001 | −0.181 | <0.001 |
| TTC21A | DC | −0.137 | 0.003 | −0.106 | 0.020 |
| TTC21A | Eosinophils | −0.044 | 0.334 | 0.009 | 0.838 |
| TTC21A | iDC | −0.101 | 0.027 | −0.049 | 0.286 |
| TTC21A | Macrophages | −0.060 | 0.187 | −0.088 | 0.053 |
| TTC21A | Mast cells | −0.042 | 0.357 | −0.018 | 0.690 |
| TTC21A | Neutrophils | −0.116 | 0.011 | −0.158 | <0.001 |
| TTC21A | NK CD56bright cells | −0.120 | 0.009 | −0.034 | 0.457 |
| TTC21A | NK CD56dim cells | −0.146 | 0.001 | −0.092 | 0.043 |
| TTC21A | NK cells | −0.025 | 0.585 | 0.084 | 0.066 |
| TTC21A | pDC | 0.050 | 0.271 | 0.082 | 0.072 |
| TTC21A | T cells | −0.098 | 0.032 | −0.113 | 0.013 |
| TTC21A | T helper cells | 0.049 | 0.286 | 0.044 | 0.341 |
| TTC21A | Tcm | 0.274 | <0.001 | 0.220 | <0.001 |
| TTC21A | Tem | 0.102 | 0.025 | 0.099 | 0.030 |
| TTC21A | Tfh | −0.008 | 0.863 | −0.004 | 0.931 |
| TTC21A | Tgd | −0.049 | 0.287 | −0.038 | 0.404 |
| TTC21A | Th1 cells | −0.142 | 0.002 | −0.158 | <0.001 |
| TTC21A | Th17 cells | 0.039 | 0.399 | 0.086 | 0.058 |
| TTC21A | Th2 cells | −0.258 | <0.001 | −0.257 | <0.001 |
| TTC21A | Treg | −0.075 | 0.100 | −0.029 | 0.526 |
DC, dendritic cells; iDC, immature DC; pDC, plasmacytoid DC; Tcm, T central memory; Tfh, T follicular helper; Tgd, T gamma delta.
Discussion
There are few studies on the TTC21A gene in the literature. And only one previous study confirmed that TTC21A is an excellent prognostic indicator of lung adenocarcinoma (28). However, our current study confirms that TTC21A expression is positively associated with poor prognosis, while TTC21B was not correlated with the prognosis in CRC.
First, we found that changes in TTC21A expression level were associated with the prognosis of CRC. Up-regulated expression of TTC21A is a freelance poor prognostic issue. At the same time, inflated TTC21A expression was unnoticeably related to a range of clinical options, as well as high tumor stage and distant metastasis. Additionally, this study found that the expression of TTC21A was related to multiple immune markers sets and immune infiltration levels in CRC. Therefore, the results of previous studies counsel that TTC21A might have a possible impact on tumor immunity and will be a prospective tumor biomarker.
This study conjointly found that TTC21A expression was correlative with prognosis in CRC patients: up-regulation of TTC21A expression advised poor prognosis. We tend to discover variations in TTC21A expression between normal and tumor tissues of CRC. To investigate the expression mechanism and relationship of TTC21A in tumors, we tend to download the DATA set of TCGA. R-3.6.3 statistical analysis showed that TTC21A expression was related to numerous clinical options, high tumor stage, and distant metastasis. Multivariate analysis also found that TTC21A expression was a freelance prognostic think about CRC patients.
A meaningful correlation between TTC21A expression and the level of immune invasion in CRC was obtained in our study. GSEA analysis showed that TTC21A expression was significantly associated with the immune infiltration levels of T cells, NK cells, and neutrophils in CRC. Similarly, the link between completely different immune cells and TTC21A expression suggests that TTC21A plays an essential role in regulating the tumor immune microenvironment in CRC. Firstly, we tend to find that the proportion of NK cells and other T cells enlarged considerably within the high expression cluster compared with the low expression cluster.
Then, we have a tendency to use ssGSEA to verify that TTC21A expression was negatively correlative with T cells. These results recommend a possible restrictive role of TTC21A within the abundance of tumor-associated T cells. Th1, Th2, Tem, and Th17 square measure T cells with entirely different functions. As with most solid tumors, CRC has immune cell infiltrations that influence completely different outcomes: Th1, Th2, Tem, and Th17 square measure T cells with entirely different functions. Like most solid tumors, CRC has immune cell infiltrates that influence outcome: infiltration of Th1 and CD8+ T cells was related to an honest prognosis, whereas infiltration of Th17 cells promoted tumor-genesis and was related to reduced disease-free survival in CRC patients (32). These associations might recommend a potential mechanism by that TTC21A regulates lymph cell performance in CRC. These results recommend that TTC21A plays a significant role in regulation and recruiting immune infiltrating cells in CRC.
Research on the role of TIICs in human tumors has generally focused on T cells, and there have been several reports on their response to immune checkpoint inhibitors (33-36). This study appends the evolving literature examining T-cells as a favorable prognostic aspect. Therefore, the effect of TTC21A on the poor prognosis of CRC is consistent with the effect of higher T cell and NK cell abundance, suggesting a possible mechanism by which TTC21A influences the overall survival of CRC.
The limitations of our study were as follows: firstly, due to the lack of experiments, our results cannot be verified. Secondly, the data used in this study came from public databases, and the information quality of these data cannot be assessed. Thirdly, the sample size of the data concerned was tiny. Also, the study didn’t cover different races and regions, which can affect gene expression in CRC. All in all, since our study solely targeted genes usually known as considerably dynamic in TCGA data sets, age, tumor classification, stage, and alternative characteristics weren’t thought-about very well. Therefore, some biological information could also be unnoted in our study.
Therefore, exaggerated expression of TTC21A correlates to the poor prognosis of CRC. Moreover, different changes in TTC21A expression were related to different proportions of immune cells like T cells, neutrophils, and NK cells in CRC. Finally, TTC21A will considerably have an effect on immune infiltration and should be used as a prognostic biomarker of CRC.
Acknowledgments
Funding: This work was supported by a grant from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2674/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-21-2674/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Silva-Fisher JM, Dang HX, White NM, et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun 2020;11:2156. [Crossref] [PubMed]
- Li J, Huang L, Zhao H, et al. The Role of Interleukins in Colorectal Cancer. Int J Biol Sci 2020;16:2323-39. [Crossref] [PubMed]
- Wu X, Wu Y, He L, et al. Effects of the intestinal microbial metabolite butyrate on the development of colorectal cancer. J Cancer 2018;9:2510-7. [Crossref] [PubMed]
- Cao M, Li H, Sun D, et al. Cancer burden of major cancers in China: A need for sustainable actions. Cancer Commun (Lond) 2020;40:205-10. [Crossref] [PubMed]
- Chen WQ, Li H, Sun KX, et al. Report of Cancer Incidence and Mortality in China, 2014. Zhonghua Zhong Liu Za Zhi 2018;40:5-13. [PubMed]
- Teo RD, Hwang JY, Termini J, et al. Fighting Cancer with Corroles. Chem Rev 2017;117:2711-29. [Crossref] [PubMed]
- Rosato PC, Wijeyesinghe S, Stolley JM, et al. Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat Commun 2019;10:567. [Crossref] [PubMed]
- Kim OY, Park HT, Dinh NTH, et al. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat Commun 2017;8:626. [Crossref] [PubMed]
- Murciano-Goroff YR, Warner AB, Wolchok JD. The future of cancer immunotherapy: microenvironment-targeting combinations. Cell Res 2020;30:507-19. [Crossref] [PubMed]
- Huang Z, Song W, Chen X. Supramolecular Self-Assembled Nanostructures for Cancer Immunotherapy. Front Chem 2020;8:380. [Crossref] [PubMed]
- Snezhkina AV, Krasnov GS, Lipatova AV, et al. The Dysregulation of Polyamine Metabolism in Colorectal Cancer Is Associated with Overexpression of c-Myc and C/EBPβ rather than Enterotoxigenic Bacteroides fragilis Infection. Oxid Med Cell Longev 2016;2016:2353560. [Crossref] [PubMed]
- Li Q, Liu X, Wang D, et al. Prognostic value of tertiary lymphoid structure and tumour infiltrating lymphocytes in oral squamous cell carcinoma. Int J Oral Sci 2020;12:24. [Crossref] [PubMed]
- Simula L, Pacella I, Colamatteo A, et al. Drp1 Controls Effective T Cell Immune-Surveillance by Regulating T Cell Migration, Proliferation, and cMyc-Dependent Metabolic Reprogramming. Cell Rep 2018;25:3059-73.e10. [Crossref] [PubMed]
- Yang J, Xu J, E Y, et al. Predictive and prognostic value of circulating blood lymphocyte subsets in metastatic breast cancer. Cancer Med 2019;8:492-500. [Crossref] [PubMed]
- Reichling C, Taieb J, Derangere V, et al. Artificial intelligence-guided tissue analysis combined with immune infiltrate assessment predicts stage III colon cancer outcomes in PETACC08 study. Gut 2020;69:681-90. [Crossref] [PubMed]
- Disis ML, Taylor MH, Kelly K, et al. Efficacy and Safety of Avelumab for Patients With Recurrent or Refractory Ovarian Cancer: Phase 1b Results From the JAVELIN Solid Tumor Trial. JAMA Oncol 2019;5:393-401. [Crossref] [PubMed]
- Ganesan AP, Clarke J, Wood O, et al. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat Immunol 2017;18:940-50. [Crossref] [PubMed]
- Mileo AM, Nisticò P, Miccadei S. Polyphenols: Immunomodulatory and Therapeutic Implication in Colorectal Cancer. Front Immunol 2019;10:729. [Crossref] [PubMed]
- Wang W, Allard BA, Pottorf TS, et al. Genetic interaction of mammalian IFT-A paralogs regulates cilia disassembly, ciliary entry of membrane protein, Hedgehog signaling, and embryogenesis. FASEB J 2020;34:6369-81. [Crossref] [PubMed]
- Allard BA, Wang W, Pottorf TS, et al. Thm2 interacts with paralog, Thm1, and sensitizes to Hedgehog signaling in postnatal skeletogenesis. Cell Mol Life Sci 2021;78:3743-62. [Crossref] [PubMed]
- Allan RK, Ratajczak T. Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 2011;16:353-67. [Crossref] [PubMed]
- Liu W, He X, Yang S, et al. Bi-allelic Mutations in TTC21A Induce Asthenoteratospermia in Humans and Mice. Am J Hum Genet 2019;104:738-48. [Crossref] [PubMed]
- Tan XF, Chen Q, Hua SH, et al. Roles of Interferon Induced Protein with Tetratricopeptide Repeats (IFIT) Family in Cancer. Curr Med Chem 2021;28:5034-47. [Crossref] [PubMed]
- Graham JB, Canniff NP, Hebert DN. TPR-containing proteins control protein organization and homeostasis for the endoplasmic reticulum. Crit Rev Biochem Mol Biol 2019;54:103-18. [Crossref] [PubMed]
- Jiang Y, Zhang C, Zhang J, et al. Comprehensive analysis of the prognosis and biological significance for IFIT family in skin cutaneous melanoma. Int Immunopharmacol 2021;101:108344. [Crossref] [PubMed]
- Pidugu VK, Pidugu HB, Wu MM, et al. Emerging Functions of Human IFIT Proteins in Cancer. Front Mol Biosci 2019;6:148. [Crossref] [PubMed]
- Wang W, Ren S, Wang Z, et al. Increased expression of TTC21A in lung adenocarcinoma infers favorable prognosis and high immune infiltrating level. Int Immunopharmacol 2020;78:106077. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Liu J, Lichtenberg T, Hoadley KA, et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018;173:400-16.e11. [Crossref] [PubMed]
- Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol 2012;30:2678-83. [Crossref] [PubMed]
- Perez LG, Kempski J, McGee HM, et al. TGF-β signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat Commun 2020;11:2608. [Crossref] [PubMed]
- Ng SSM, Nagy BA, Jensen SM, et al. Heterodimeric IL15 Treatment Enhances Tumor Infiltration, Persistence, and Effector Functions of Adoptively Transferred Tumor-specific T Cells in the Absence of Lymphodepletion. Clin Cancer Res 2017;23:2817-30. [Crossref] [PubMed]
- Gadgeel SM, Pennell NA, Fidler MJ, et al. Phase II Study of Maintenance Pembrolizumab in Patients with Extensive-Stage Small Cell Lung Cancer (SCLC). J Thorac Oncol 2018;13:1393-9. [Crossref] [PubMed]
- Chiu DK, Tse AP, Xu IM, et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun 2017;8:517. [Crossref] [PubMed]
- Nguyen A, Ramesh A, Kumar S, et al. Granzyme B nanoreporter for early monitoring of tumor response to immunotherapy. Sci Adv 2020;6:eabc2777. [Crossref] [PubMed]



